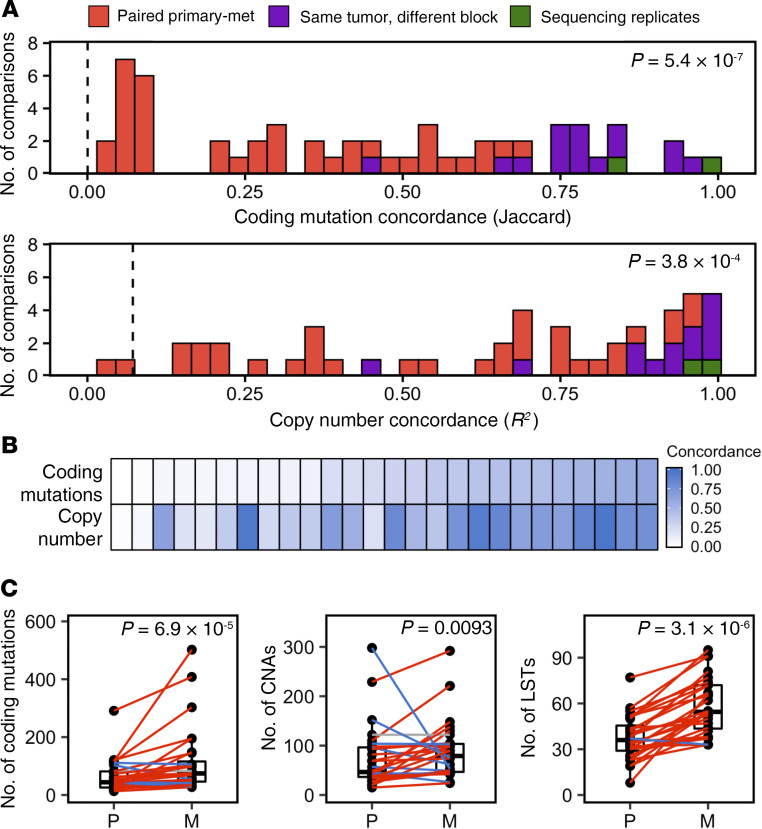
Figure 1. Paired primary-metastatic tumors are genomically distinct.
(A) Concordance for coding mutations (Jaccard index) and genome-wide copy number (R2) between primary-metastasis tumor pairs (orange, n = 28), multiple regions assayed from different tissue blocks from the same tumor (purple, n = 14), and sequencing replicates (green, n = 12). Concordance between primary-metastasis tumor pairs is substantially lower than concordance values between multiple regions within the same tumor (1-sided Wilcoxon’s rank-sum test). Dashed vertical lines indicate mean concordance between unrelated tumors. (B) Concordance determined for coding mutations and genome-wide CNs are significantly correlated, R2 = 0.47. Columns represent primary-metastasis tumor pairs. (C) Metastases (M) exhibit increased numbers of somatic coding mutations, CNAs, and LSTs compared with the primary tumors (P) from which they arose (1-sided Wilcoxon’s signed-rank test). Colored lines indicate patients for whom metastatic tumors exhibited an increased (red) or decreased (blue) number of alterations compared with primary tumors from which they arose.