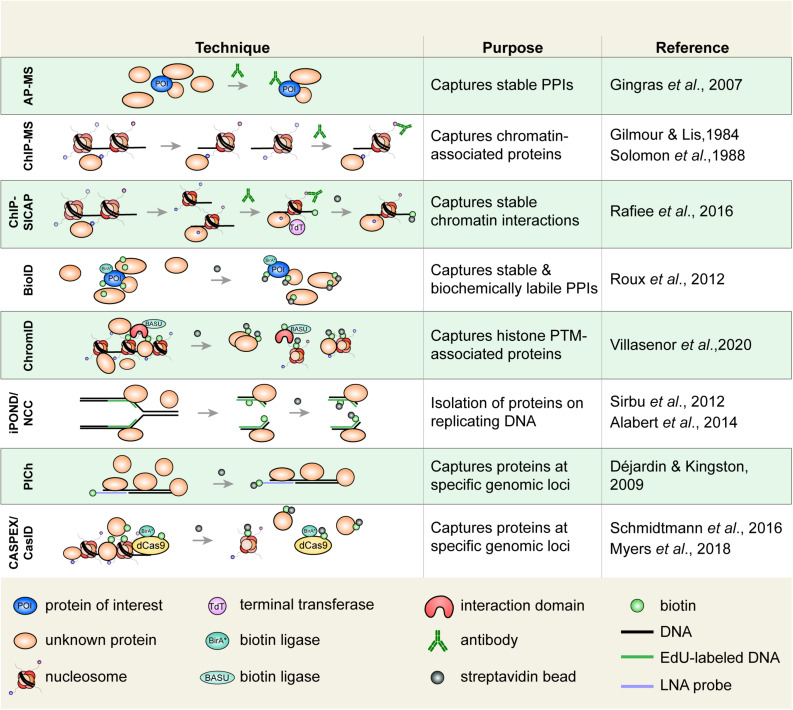
FIGURE 3.
Select techniques used to study histone or chromatin-associated proteins. Affinity purification-mass spectrometry (AP-MS) is used to isolate biochemically stable protein–protein interactions. Chromatin immunoprecipitation (ChIP), in which an epitope of interest is isolated from sheared chromatin fragments, can be coupled to MS to identify associated proteins. ChIP-SICAP uses an additional DNA biotinylation step to wash proteins not directly bound to chromatin. Proximity-dependent labeling techniques are increasingly used to capture stable and biochemically labile protein interactions. Biotin identification (BioID) uses a biotin ligase fused to a protein of interest to biotinylate proximal proteins. Biotinylated proteins are captured on streptavidin beads. ChromID is similar to BioID in that a biotin ligase, BASU, is fused to a histone-binding domain to biotinylate proteins near a PTM of interest. Isolation of proteins on nascent DNA (iPOND)/nascent chromatin capture (NCC), are used to isolate proteins associated with replicating DNA. Cells are pulsed with a thymidine analog (e.g., EdU), which is incorporated on replicated DNA enabling the isolation of replicated chromatin fragments. Proteomics of isolated chromatin (PICh) is used to identify proteins that are bound to a specific genomic region. A biotin-tagged locked nucleic acid (LNA) probe is used to isolate chromatin fragments with DNA complementary to the probe. In CASPEX/CasID, catalytically dead Cas9 (dCas9) is fused to APEX or BirA*, respectively, to perform biotin labeling of a specific genomic locus. PPIs, protein–protein interactions.