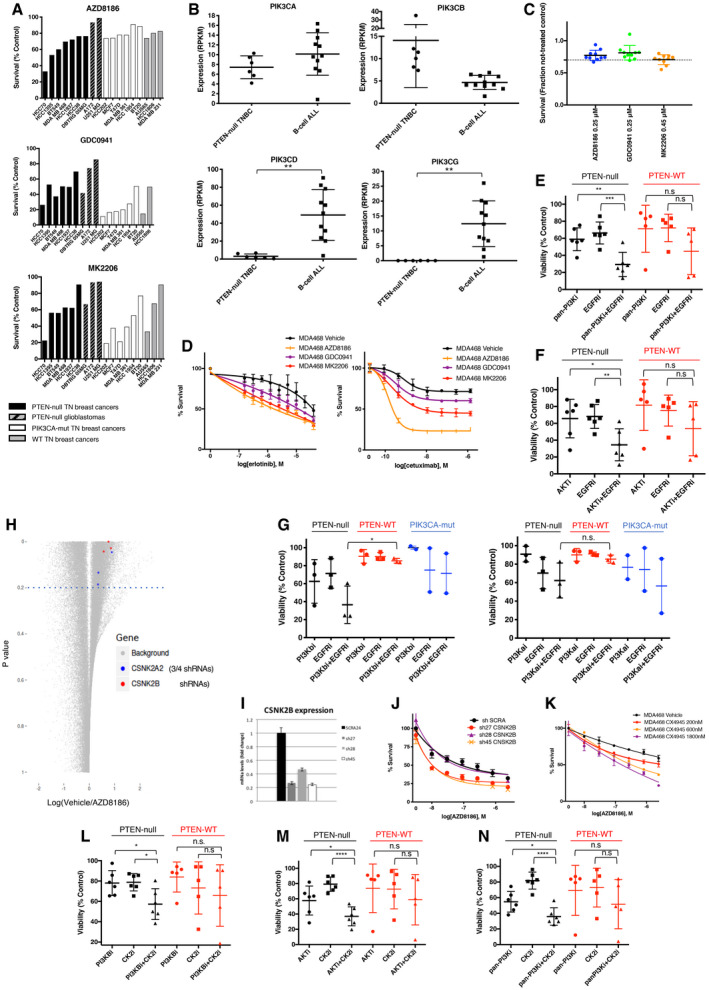
Figure EV1. A short‐hairpin screening identified hits whose inhibition enhanced the effect of PI3K pathway inhibitors on PTEN‐deficient triple‐negative breast cancers. Related to Fig 1 .
-
ACell viability of cell lines treated for 4 days with 0.25 μM AZD8186, 1 μM GDC0941, or 1 μM MK2206. PTEN‐deficient triple‐negative breast cancer cell lines are represented by black bars, PTEN‐deficient glioblastoma cell lines by black/gray dashed black, PIK3CA‐mutant breast cancer cell lines by white bars, and PIK3CA WT and PTEN WT breast cancer cell lines by gray bars. Each bar represents the mean of three experiments.
-
BPlot of expression data extrapolated from RPKM RNAseq files of Broad Institute Cancer Cell Line Encyclopedia (CCLE, https://portals.broadinstitute.org/ccle), and specifically showing expression of PIK3CA, PIK3CB, PIK3CD, and PIK3CG across PTEN‐null TNBC cell lines (namely: MDA‐MB‐468, HCC1395, HCC1937, HCC38, HCC70, and BT‐549) and B‐cell acute lymphocytic leukemia (B‐cell ALL) lines (namely: BDCM, JM1, KOPN8, MHHCALL3, MHHCALL4, MUTZ5, NALM6, REH, RS411, SEM, and SUPB15). Mean ± SD. Statistical significance of two‐tailed unpaired t‐test in PIK3CD expression **P = 0.0014 and in PIK3CG expression **P = 0.0015.
-
CCell viability of MDA‐MB‐468 cells infected with 10 shRNAs pools during the shRNA screen (the library was divided into 10 pools, and each dot represents one pool) and co‐treated as indicated. Viability of cells was normalized to cell numbers in the vehicle‐treated condition within each pool and expressed as mean ± SD.
-
DMDA‐MB‐468 was treated with serial dilutions of erlotinib or cetuximab in the presence of vehicle, AZD8186 (0.25 μM), GDC0941 (0.25 μM), or MK2206 (0.45 μM), as indicated. Cell viability was measured after 4 days (erlotinib) or 6 days (cetuximab) and normalized within each of the PI3K pathway inhibitor‐treated condition to the viability in the absence of erlotinib or cetuximab. Average ± SD of triplicates and representative of two independent experiments.
-
E, FViability of six PTEN‐null vs five PTEN‐WT TNBC cell lines treated for 6 days with pan‐PI3Ki (GDC0941 1 μM) (E), or AKTi (MK2206 0.45 μM) (F), alone or in combination with EGFRi (gefitinib 3 μM). Mean of three independent experiments ± SD. Statistical significance of two‐tailed unpaired Student's t‐test in PTEN‐null pan‐PI3Ki vs pan‐PI3Ki + EGFRi **P = 0.0041, PTEN‐null EGFRi vs pan‐PI3Ki + EGFRi ***P = 0.0008, PTEN‐WT pan‐PI3Ki vs pan‐PI3Ki + EGFRi n.s. P = 0.166, PTEN‐WT EGFRi vs pan‐PI3Ki + EGFRi n.s. P = 0.0897, PTEN‐null AKTi vs AKTi + EGFRi *P = 0.0281, PTEN‐null EGFRi vs AKTi + EGFRi ***P = 0.0059, PTEN‐WT AKTi vs AKTi + EGFRi n.s. P = 0.1936, PTEN‐WT EGFRi vs AKTi + EGFRi n.s. P = 0.2302.
-
GViability of three PTEN‐null (MDA‐MB‐468, HCC70 and HCC1937), three PTEN‐WT (MDA‐MB‐231, MDA‐MB‐157, and HCC1428) and two PIK3CA‐mutant (SUM159 and BT20) TNBC cell lines treated for 6 days with PI3Kβi (AZD8186 90 nM) (left), or PI3Kαi (BYL719 1.2 μM) (right), alone or in combination with EGFRi (gefitinib 3 μM). Mean of 3–4 independent experiments ± SD. Statistical significance of two‐tailed unpaired Student's t‐test PI3Kbi + EGFR treatments in PTEN‐null vs PTEN‐WT *P = 0.0158, PI3Kai + EGFR treatments in PTEN‐null vs PTEN‐WT n.s. P = 0.1081.
-
HDot‐plot showing the fold change (log2) in number of reads between vehicle and AZD8186‐treated conditions vs the P‐value of the difference between the two treatment conditions for each shRNA. Three out of four shRNAs targeting CSNK2A2 and three out of seven shRNAs targeting CSNK2B showed a P‐value < 0.2 calculated by two‐sided paired t‐test and are highlighted in the figure.
-
I, JMDA‐MB‐468 was infected with the indicated shRNAs targeting CSNK2B and selected by puromycin. CSNK2B mRNA was then measured by RT–qPCR (I), and cell viability was measured after 4 days of treatment with serial dilutions of AZD8186 (J) Average ± SD of triplicates and representative of three independent experiments.
-
KMDA‐MB‐468 cells were treated with serial dilutions of AZD8186 in combination with vehicle or with the indicated concentrations of CX4945. Viability was measured after 6 days and normalized within each of the CX4945‐treated condition to the viability in the absence of AZD8186. Mean ± SD of triplicates and representative of three independent experiments.
-
L–NViability of six PTEN‐null vs five PTEN‐WT TNBC cell lines treated for 6 days with PI3Kβi (AZD8186 10 nM) (L), AKTi (MK2206 0.45 μM) (M), or pan‐PI3Ki (GDC0941 1 μM) (N) alone or in combination with CK2i (CX4945 1 μM). Mean of 3–5 independent experiments ± SD. Statistical significance calculated by two‐tailed unpaired Student's t‐test. PI3KBi vs PI3KBi + CK2i *P = 0.0265, CK2i vs PI3KBi + CK2i *P = 0.0128; AKTi vs AKTi + CK2i *P = 0.0494, CK2i vs AKTi + CK2i ****P < 0.0001; pan‐PI3Ki vs pan‐PI3Ki + CK2i *P = 0.0235, CK2i vs pan‐PI3Ki + CK2i ****P < 0.0001; n.s. P > 0.05.
