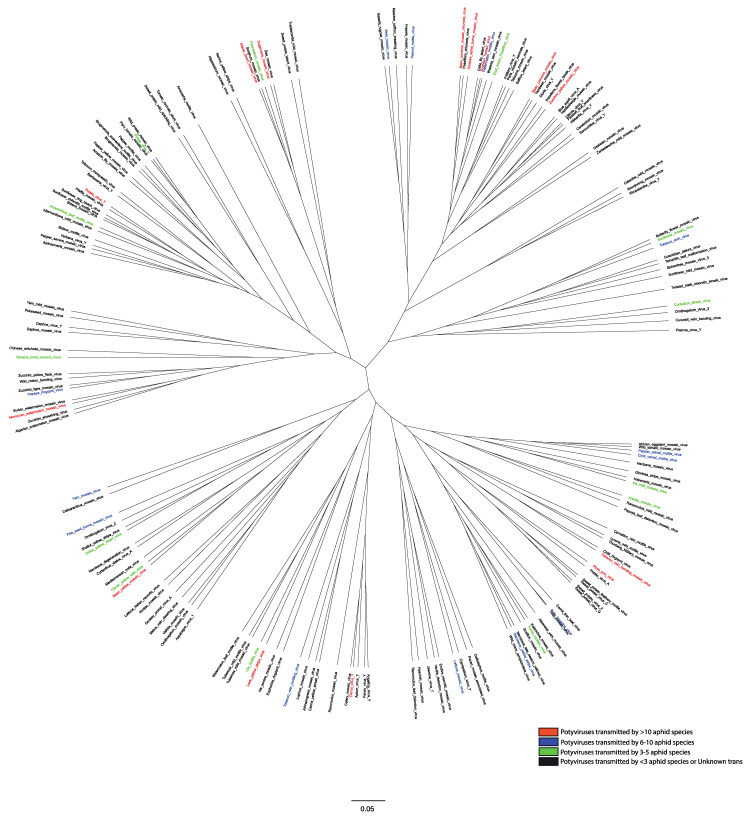
Figure 3.
Phylogenetic analysis based on the coat protein (CP) sequences of 176 potyviruses. The genomic sequence for each exemplar potyvirus species was downloaded through NCBI GenBank. Accession numbers are provided in Supplementary Table S1. Nucleotide sequences were aligned using MAFFT v7.127b [37] and then subalignments for the CP coding regions were then manually refined by deleting highly gapped and ambiguously aligned sites. Phylogenetic trees for CP gene were then reconstructed in BEAST v 2.5.2 [38]. Sequences were assumed to evolve under a Hasegawa–Kishono–Yano (HKY) substitution model with gamma rate heterogeneity across sites. A strict molecular clock with a fixed substitution rate of 1.0 per unit time was assumed such that the resulting phylogenies have branch lengths given in units of substitutions per site. BEAST was run for 10 million Markov chain Monte Carlo (MCMC) iterations, sampling a tree from the posterior distribution every 10,000 iterations. The posterior distribution of trees was summarized as a Maximum Clade Credibility (MCC) tree with mean node heights. The resulting MCC tree for CP was then visualized in FigTree and uncertainty in the tree topology was assessed using the posterior support for each split (clade) in the tree. The potyvirus species in the tree was color-coded on the basis of number of aphid vectors from published scientific studies (Supplementary Table S1).