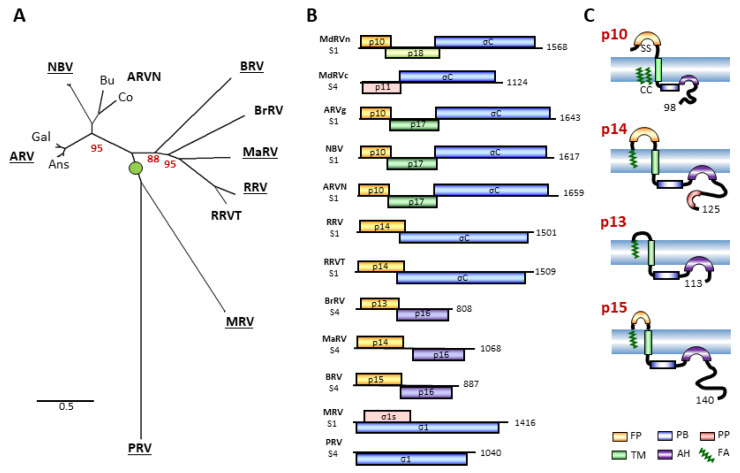
Figure 1.
The diversity of orthoreovirus species, their polycistronic genome segments, and their fusion-associated small transmembrane (FAST) proteins. (A) Maximum likelihood unrooted phylogram based on 23 concatenated amino acid sequences of the seven consistently homologous structural proteins (core shell, core RdRp, core turret, core NTPase, core clamp, outer-shell, outer-clamp) encoded by the indicated 10 species or proposed species of orthoreoviruses (in boldface). See Supplementary Table S1 for a list of viruses, proteins, and accession numbers. Lowercase labels denote isolates within a species: Gal and Ans are landfowl and waterfowl isolates of avian orthoreovirus (ARV); Bu and Co are bulbul and corvid isolates of neoavian orthoreovirus (ARVN). Bootstrap values (500 replicates) are shown for branch points where the associated taxa clustered together in less than 100% of the replicate trees. The tree with the highest log likelihood is shown. Branch lengths are to scale and measured by the number of substitutions per site. (B) Diagrams of the polycistronic S1 and S4 genome segments from the indicated orthoreovirus species, drawn to approximate scale. Open reading frames are depicted by rectangles, labeled to indicate the protein product and color-coded to depict functional or evolutionary protein relationships: yellow—FAST protein; blue—fiber protein; green—potential cell cycle inhibitory protein; purple—homologous proteins with no defined function; pink—nucleocytoplasmic shuttling protein. Numbers refer to mRNA length. (C) Diagrams of the orthoreovirus FAST proteins, depicting their topology in the plasma membrane. Structural motifs contained within their N-terminal ectodomains and C-terminal cytoplasmic endodomains are color-coded as described in the legend at the bottom. Numbers indicate the number of residues in each protein.