Fig. 7.
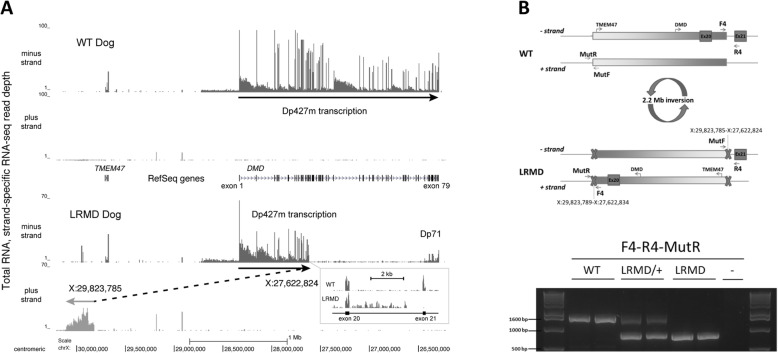
RNA-seq-based identification of the LRMD mutation. a A 4 Mb region spanning canFam3 chrX:30,200,000–26,200,000 is shown with RNA-seq read depths observed in a biceps femoris biopsy experiment plotted with RefSeq gene annotations. The upper panel shows Dp427m minus-strand transcript levels (DMD exons 1 through 79) seen with an unaffected dog, while the lower panel shows the inversion inferred from minus-strand DMD transcription and plus-strand ectopic transcription seen with LRMD8. The boxed inset shows a detailed view of intron 20 transcription from these two animals. b Schematic representation of the LRMD mutation, and LRMD genetic diagnosis by PCR. Two pairs of primers were used: the first set (F4R4) amplifies a 1661 bp product in the WT dog but not in the LRMD dog (breakpoint between these two primers). A second pair of primers (MutFMutR) was designed to amplify the distant site across the breakpoint yielding a 1939 bp product in the WT sample but not in the LRMD (data not shown). The proposed diagnostic PCR test relies on the use of F4 as a forward primer, and the combination of R4 and MutR as reverse primers to amplify either the WT or the LRMD allele. Using this test, a unique PCR product around 1600 bp was observed in the samples obtained from the LRMD healthy littermates (amplicon including F4-R4); a unique PCR product around 900 bp was observed in the samples of the two affected LRMD dogs (amplicon including F4-MutR); and both amplicons were observed in samples obtained from carrier females