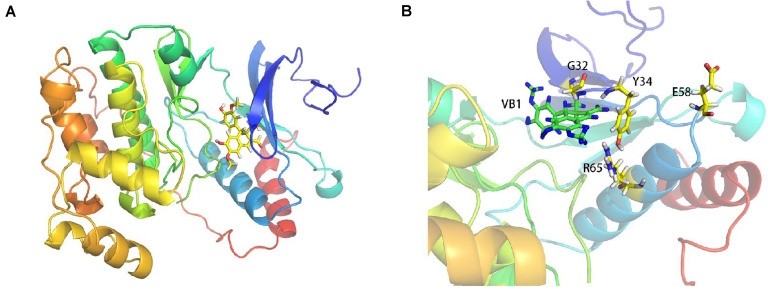
FIGURE 3.
The possible target sites of MAPK1 binding VB1 are predicted by computer-aided methods including molecular docking and molecular dynamics simulation. The MAPK1-VB1 binding site was predicted by Discovery Studio 2.5 and Autodock Vina. The molecular docking study and the Molecular dynamics (MD) simulations were performed to explore the binding details base the docking results. (A) The molecular docking and molecular dynamics simulation results showed VB1 could directly bind MAPK1 protein. (B) The five amino acid residues (G32, Y34, K46, E58, and R65) of MAPK1 were predicted and K338 residue was another possible key residue through computer-aid calculation (data not shown).