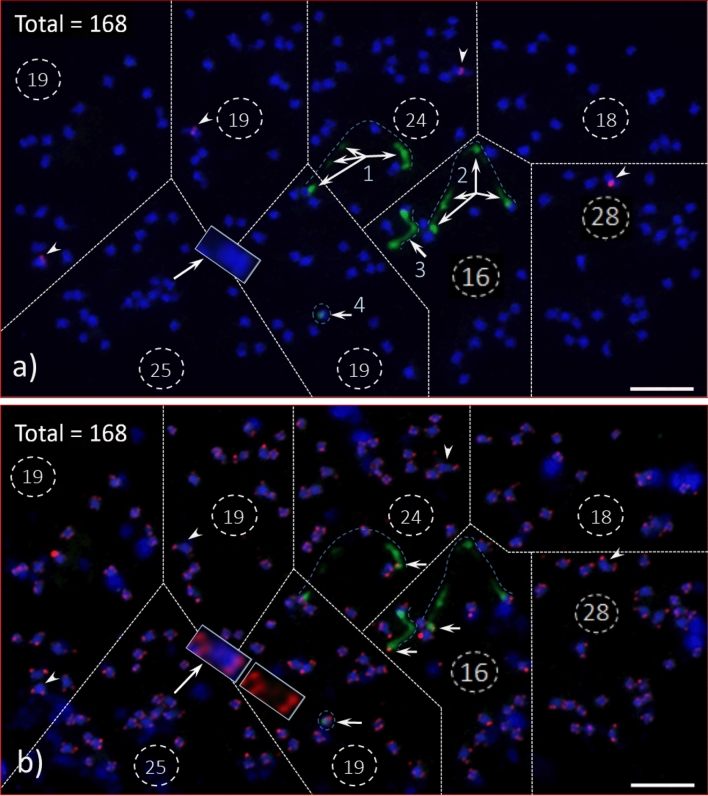
Figure 7.
Adansonia digitata L. (Seedling-2) pro-metaphase chromosome spread analyzed with 45S and 5S rDNA probes (1st hybridization, “a”) and re-hybridized with 45S rDNA (as a control) and ATRS probes (2nd hybridization or re-hybridized, “b”) a) As many as thirteen 45S rDNA (green signals, arrows) from four NOR bearing chromosomes (dotted lines over green signals numbered as 1, 2, 3 and 4) and four 5S rDNA (red, arrowheads) signals, the spread is divided into eight sections to facilitate accurate counting of chromosomes and the total number is 168 (19 + 19 + 24 + 18 + 25 + 19 + 16 + 28 = 168). One large metacentric chromosome (see insert in the bottom left-hand side section) may appear as two chromosomes (due to its sharp constriction in the center of the chromosome). In such scenario, (a) ATRS hybridization (see insert in “b”) would be the option to confirm a single chromosome; (b) The same cell was re-hybridized (i.e., 2nd hybridization, see Method for details) with 45S rDNA (as control, green signal) ATRS probes (red signals) after washing off probes from the 1st hybridization. The 5S rDNA signals are missing (arrowheads), which confirmed the 1st hybridization probes were completely washed off. Though the terminal end of each 45S rDNA-bearing chromosomes showed no visible sign of satellites, the telomere signals confirmed (arrows) that these chromosomes are protected by telomere repeat DNA sequences. The same large metacentric chromosome (insert in “a”) showed a pair of ATRS signals (red) at each end (inserts in “b”) and no signals in the middle of the chromosome that further confirmed as one chromosome, and the total count was 168. Scale bars are 5 µm.