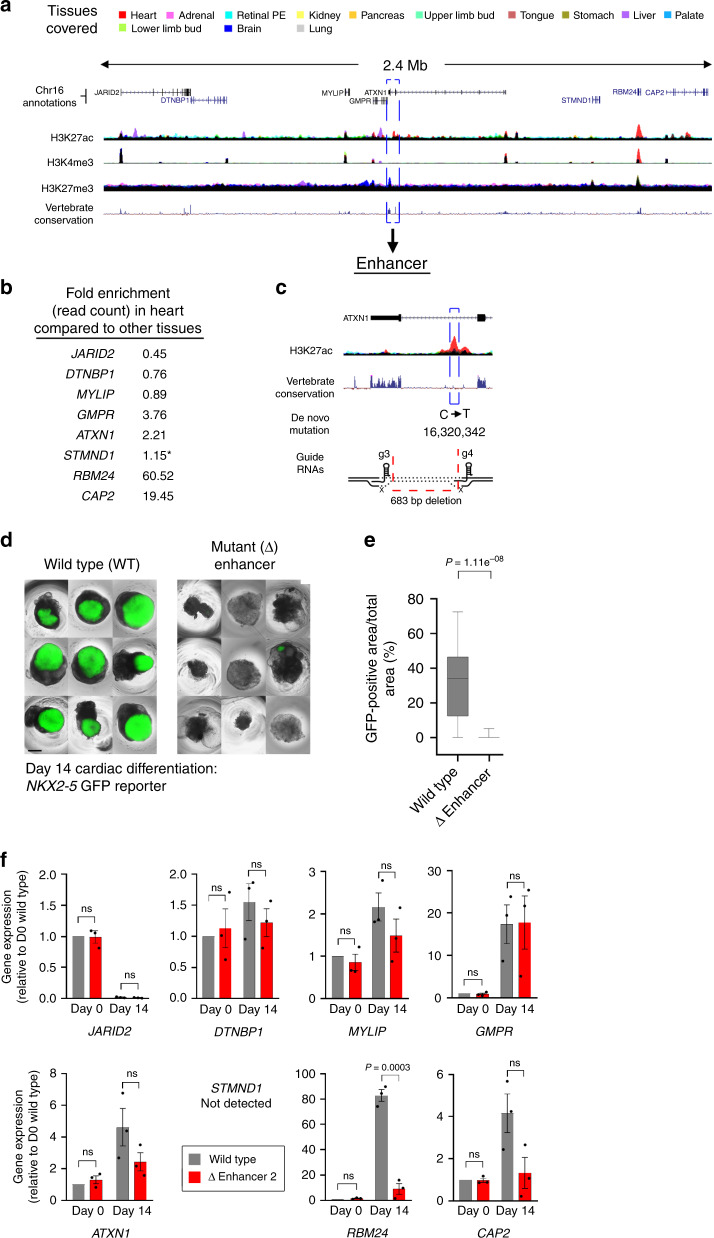
Fig. 8. Deletion of enhancer upstream of RBM24 disrupts cardiomyocyte differentiation.
a Schematic of a 2.4 mb locus on chromosome 6 containing eight protein-coding genes centred on a cardiac-specific H3K27ac peak (red, broken line box) within the last intron of ATXN1. This enhancer harbours a DNM from the DDD cohort associated with congenital heart disease2. The histone modification tracks contain datasets from all the colour-coded tissues. b Fold enrichment in the heart/LV dataset compared with the average across all other tissues for RNAseq read counts of the genes shown in (a). RBM24 and CAP2 are considerably enriched in the human embryonic heart. c Magnified schematic of the enhancer shown in (a) showing the location of the DNM and the CRISPR-Cas9 approach for deletion. d EBs from wild-type and enhancer deletion (mutant) hPSCs containing the NKX2-5-GFP reporter. The images showing nine EBs for wild-type and mutant are after 14 days of the cardiomyocyte differentiation protocol34,35. Size bar, 500 μm. e Box and whisker plot (box showing 25th−75th percentile and median line with min–max as whiskers) quantifying GFP across all wild-type (n = 29) and mutant EBs (n = 30). f RT-qPCR for expression of all the protein-coding genes across the 2.4 mb locus depicted in (a). Ten different clones were used in three independent experiments for mutant EBs with ten, ten and nine clones for wild-type control. Error bars represent S.E.M. from the three independent differentiation experiments (the individual dots). Time points for RT-qPCR were day 0 (undifferentiated hPSCs) and day 14 (cardiomyocyte progenitors34,35). While CAP2 expression appeared reduced, RBM24 was the only gene with significantly lowered expression following deletion of the cardiac-specific enhancer shown in a). Significance was assessed using a two-tailed Student’s t test (ns, not significant).