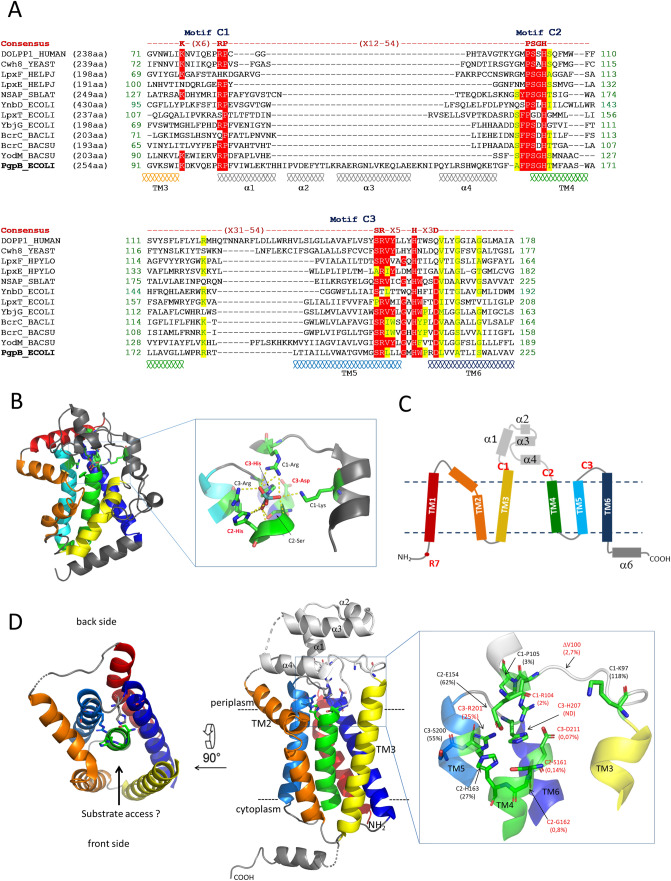
Figure 1.
(A) Sequence alignment of PAP2 consensus regions of PgpB, YbjG, LpxT and YnbD from E. coli, YodM and BcrC from B. subtilis or B. licheniformis, dolichyl-pyrophosphate phosphatases from Human (DOLPP1) and Saccharomyces cerevisiae (Cwh8), lipid A phosphatases LpxE and LpxF from H. pylori and NSAP from S. blattae. Identical and similar residues are indicated on a red and a yellow background, respectively. (B) Cartoon representation of molybdate-bound NSAP structure (PDB 1EOI) with the core helix bundle represented in rainbow color gradient from the N-terminus in blue to the C-terminus in red. (C) Topology diagram of PgpB. (D) Cartoon representation of PgpB structure (PDB 4PX7). The signature residues are labeled in red or black depending on whether or not their mutation affects the protein activity in vivo, respectively, and the residual C55-PP phosphatase activity of each variant is shown in parentheses. The figure was generated with PyMol.