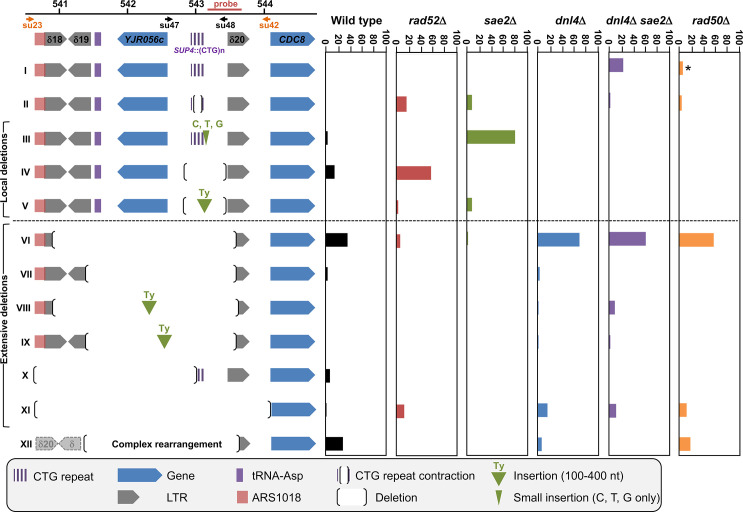
Fig 4. Summary of chromosomal rearrangements observed in wild-type and mutant strains, following SpCas9 induction.
Left: The twelve different possible outcomes following SpCas9 induction are shown, subdivided in local and extensive deletions (see text for details). The SUP4 locus is pictured and shows the position of each genetic element on yeast chromosome X. The probe used on Southern blots is shown, as well as both primer couples used to amplify the locus. In order to assess a given clone to a rearrangement type, the following rules were followed: i) when a band was detected by Southern blot, primers su47 and su48 were used to amplify the locus and sequence it. These events corresponded to types I-V. The absence of a PCR product indicated that primer su48 genomic sequence was probably deleted and therefore primers su23 and su42 were used to amplify and sequence the locus. These were classified as types IV-V events; ii) when no band was detected by Southern blot, primers su 23 and su42 were directly used to amplify and sequence the locus. These events were classified as types VI-X and XII. When no PCR product was obtained, it meant that at least one of the two primers genomic sequence was probably deleted and these events were classified as type XI. Note that this last category may also contain rare -but possible- chromosomal translocations that ended up in puting each primer in a separate chromosome, making unobtainable the PCR product. The extent of type XI deletions cannot go downstream the su42 primer, since the CDC8 gene is essential. Right: The proportion of each type or event recovered is represented for wild type and mutants. The asterisk near Type I events in the rad50Δ strain indicates that some of them were small expansions (see text). Altogether, 262 surviving clones were sequenced, distributed as follows: WT: 51, rad52Δ: 29, dnl4Δ: 61, sae2Δ: 32, dnl4Δ sae2Δ: 47, rad50Δ: 42.