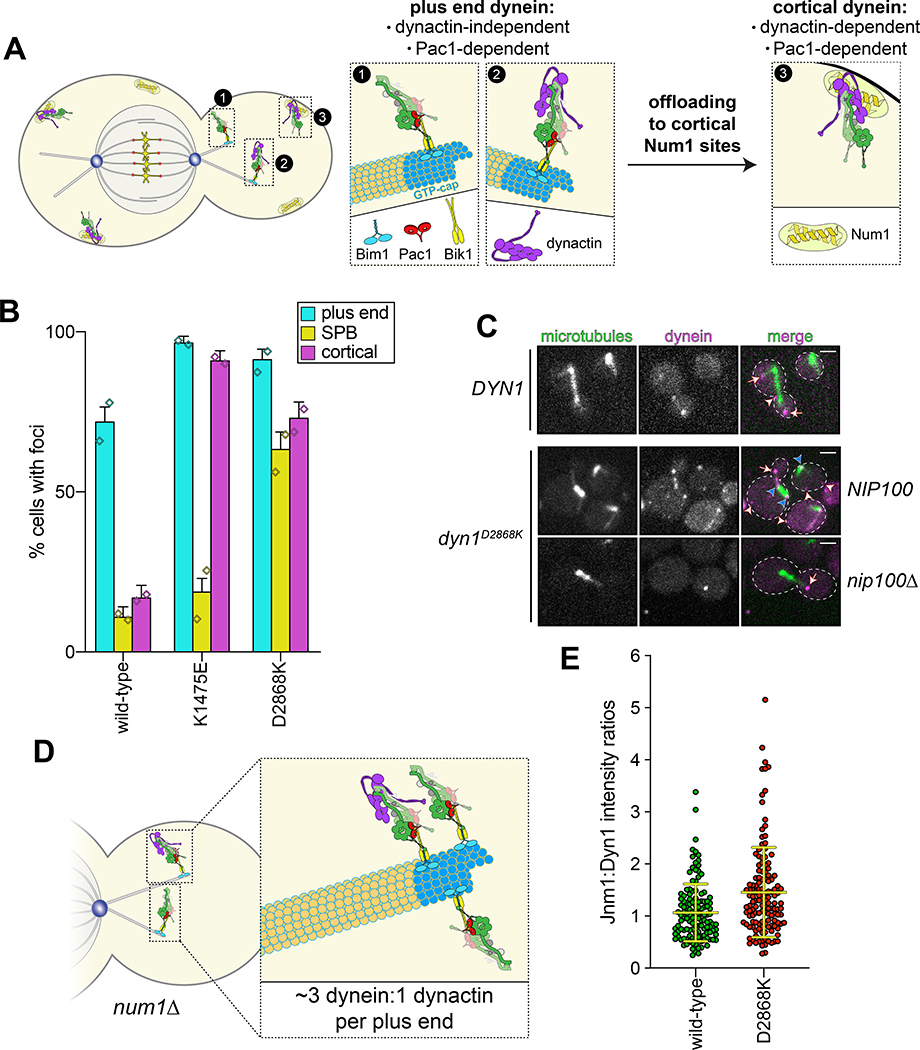
Figure 3. The autoinhibited conformation restricts cellular localization of dynein and its interaction with dynactin.
(A) Cartoon depicting the two main sites of dynein localization (microtubule plus ends, and cell cortex), and the molecular requirements for each. Dynein plus end localization (1) requires Bik150 and Pac144, with Bim1 potentially playing some role in this process, but does not require dynactin41. Rather, dynactin plus end localization (2) relies on dynein41. Subsequent to plus end targeting, dynein-dynactin complexes are offloaded to cortical Num1 sites29,61 (3). (B) Plot depicting the fraction of cells (weighted mean) with indicated mutant or wild-type Dyn1–3GFP foci cells (n = 100, 90, 82 mitotic cells from two independent experiments; error bars indicate weighted standard error of proportion; diamonds represent mean values obtained from each independent replicate experiment). (C) Representative images of wild-type or mutant dynein (D2868K) localizing in otherwise wild-type or nip100Δ (dynactin component) cells. Note the lack of cortical localization of dyneinD2868K in nip100Δ cells (white arrowheads, cortical foci; white arrows, plus end foci; blue arrowheads, SPB foci; similar results were obtained from 2 independent replicates; scale bars, 2 μm). (D) Cartoon depicting the relative ratio of dynein to dynactin at microtubule plus ends (~3 dynein:1 dynactin), based on previous quantitative ratiometric imaging49. (E) Scatter plot (shown with bars depicting mean and standard deviation values) of the ratios of fluorescence intensity values for Jnm1–3mCherry (dynactin component p50/dynamitin):Dyn1–3GFP at individual microtubule plus ends and SPBs (n = 132 and 145 plus ends or SPBs, left to right, from two independent experiments). Background corrected intensity values of colocalizing Jnm1–3mCherry and Dyn1–3GFP foci were each divided to obtain individual ratio values. Measurements were taken from num1Δ cells to prevent offloading of assembled dynein-dynactin complexes from plus ends.