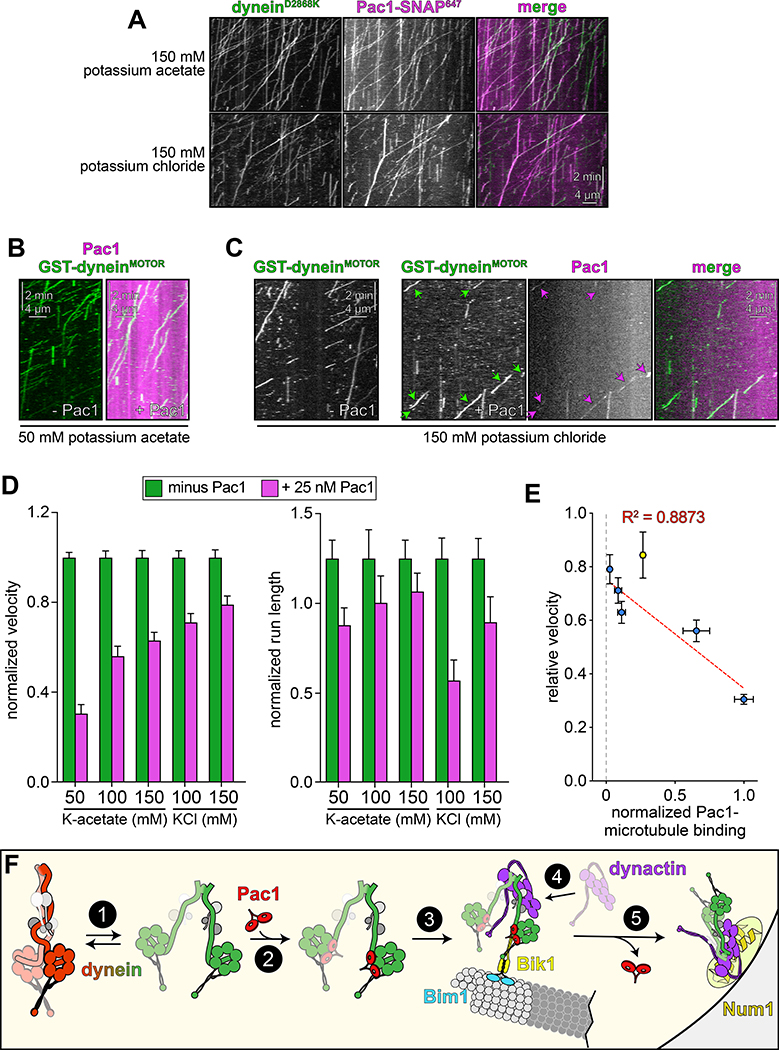
Figure 6. Reducing Pac1-microtubule binding minimizes Pac1-mediated dynein velocity reduction.
(A) Representative kymographs depicting comigrating dyneinD2868K-Pac1 complexes in motility buffers with increased ionic strength (from 3 and 2 independent replicates, top to bottom). Note that Pac1 and dynein still interact robustly in these conditions, as apparent from the high degree of persistent colocalization. (B and C) Representative kymographs depicting different motility characteristics of GST-dyneinMOTOR (used extensively in previous Pac1 studies26–28) in the presence of Pac1 when the latter is either extensively bound to the microtubule (B), or to a much less extent (C; see panel D for n values). (D) Mean normalized motility parameters (see Extended Data Fig. 7E – G for means from independent replicates, and scatter plots) of GST-dyneinMOTOR in the absence (green) or presence (magenta) of 25 nM Pac1 (dimer concentration; n = 348, 268, 396, 226, 447, 359, 385, 315, 251, 320 motors, from two independent experiments each; left to right). Error bars indicate standard error. (E) Relative degree of Pac1-microtubule binding (mean values normalized to 1; see Extended Data Fig. 6B and E for scatter plot of intensity values, and n values for each) versus mean relative velocity of GST-dyneinMOTOR in the presence of Pac1 (mean GST-dyneinMOTOR velocity in the absence of Pac1 equals 1; see panel D, and Extended Data Figs. 6F and G, and 7E – G for relative and absolute velocity values, and n values). Blue and yellow points (error bars represent standard error) are from increasing ionic strength buffer experiment (see panel D), and the cell extract experiment (see Extended Data Fig. 6D – G), respectively. The blue points were fit to a linear regression with R2 value shown. (F) Model for dynein and Pac1 activity in cells: (1) dynein stochastically switches between closed and open states, the latter of which is stabilized by Pac1 binding (2); dynein-Pac1 associates with plus ends via direct interactions with Bik1 (3), which may rely partly on Bim1. (4) Plus end dynein-Pac1 associates with dynactin, which is then offloaded to cortical Num1 (5). Given the lack of apparent Pac1 cortical foci, Pac1 likely dissociates either concomitant with, or subsequent to dynein-dynactin offloading. Also see Extended Data Figure 7.