Figure 6. Proposed working models for differential deadenylation profiles in LARP4 KO and WT MEFs (derived from Figure 3A and select panels of 5C).
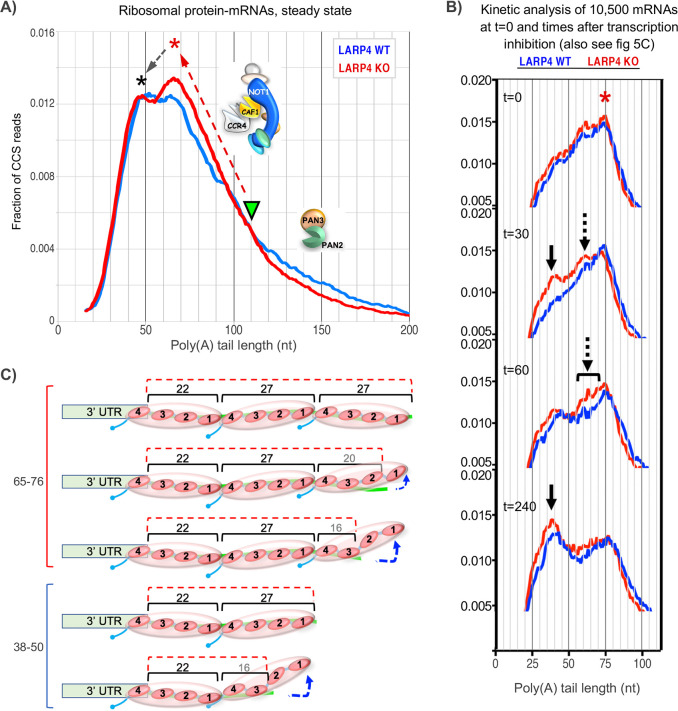
(A) LARP4 effects on phasing of a set of ~80 ribosomal protein mRNAs at steady state was revealed by plotting the fraction of CCS reads vs. PAT lengths. A green triangle points to where the KO and WT transitions from long to short PATs intersect, at ~110 nt. The upward red dashed arrow denotes a steeper incline in KO as compared to WT of long to short PAT length transition. The red asterisk denotes the ~60–75 nt PATs whose peak is comprised of a substantially higher fraction of reads in KO as compared to WT cells. The black dashed arrow denotes conversion of longer to shorter PAT complexes in the peaks denoted by the corresponding asterisks. In this model, the peak denoted by the red asterisk would correspond to PATs bound by three PABPs that include peeling off/dissociation intermediates schematized in the upper part of panel C, and the peak denoted the black asterisk would correspond to PATs bound by two PABPs. As the two profiles transition from long to shorter PATs they begin to overlap at PAT lengths of ~110 nt which would correspond to mRNP complexes containing four PABPs or two PABP-dimers. These profiles suggest that as PATs shorten from this transition point and the distal PABP would begin to dissociate, CNOT may be less impeded in the absence of LARP4 (see text). Thus LARP4 may exert a stabilizing effect at this point. (B) Transcriptome-wide dynamic PAT phasing observed in SM-PAT-seq profiles in Figure 5C illustrating a deadenylation time course after transcription inhibition by actinomycin D. Red asterisk denotes the 75 nt PAT peak at t = 0; panels depicting t = 30, 60 and 240 min are also shown as labeled. Solid black arrow denotes a ~ 38 nt PAT peak, dashed arrows and bracket denote proposed intermediate (see text and Figure 5C). (C) Schematized depiction of potential PAT-PABP(3) and PAT-PABP(2) complexes and intermediate binding forms thereof that might comprise the mRNA-PAT profile peaks in A and B, in various states of peeling off modes of PABP dissociation, modeled after Webster et al., 2018 and discussed in the text.