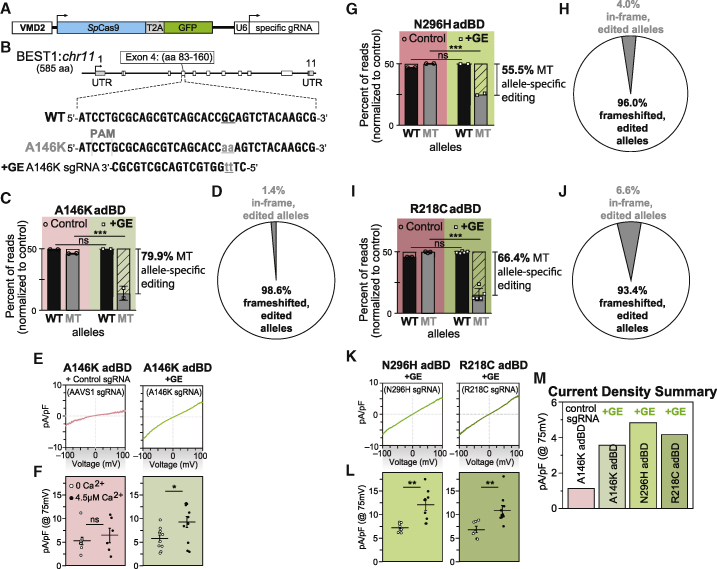
Figure 4.
Gene Editing Specifically and Efficiently Introduces Frameshifts within the Mutant Allele in adBD iPSC-RPE and Rescues CaCC Activity
(A) Lentiviral genome editing construct used to express spCas9 and mutant allele-targeted sgRNAs.
(B) Diagram showing the heterozygous base pair substitutions in Ala146Lys adBD and the design of the A146K sgRNA. The WT allele is shown above, while the mutant allele encoding p.Ala146Lys (c.436_437delinsAA) is shown below, with the mutated bases indicated in lowercase and underlined.
(C) Percentage of WT and mutant (MT; unedited and edited) allele sequencing reads in Ala146Lys iPSC-RPE treated with A146K sgRNA lentiviral genome editor (“+GE”), respectively, normalized to control (“Control,” genome edited with safe harbor AAVS1-targeting sgRNA).
(D) Indel frameshift and in-frame frequency for mutant allele-edited reads from Ala146Lys adBD iPSC-RPE (corresponds to (C)).
(E and F) (E) CaCC current density-voltage plots and (F) CaCC conductance for individual iPSC-RPE cells from single-cell patch clamp experiments for Ala146Lys iPSC-RPE treated with control (AAVS1) or mutant allele-targeted sgRNA lentiviral genome editor.
(G–J) Percentage of WT and mutant (MT; unedited and edited) allele sequencing reads in Asn296His (G) or Arg218Cys (I) adBD iPSC-RPE treated with N296H or R218C sgRNA lentiviral genome editor, respectively, normalized to control (AAVS1 sgRNA). Indel frameshift and in-frame frequency in Asn296His (H) or Arg218Cys (J) adBD iPSC-RPE treated with N296H or R218C sgRNA lentiviral genome editor, respectively (correspond to (G) and (I), respectively).
(K and L) (K) CaCC current density-voltage plots and (L) CaCC conductance for individual iPSC-RPE cells from single-cell patch clamp experiments for Asn296His or Arg218Cys adBD iPSC-RPE treated with corresponding mutant allele-targeted sgRNA lentiviral genome editor.
(M) Mean CaCC conductance at 75 mV for each adBD iPSC-RPE model. The number of cells is the same as (E) and (K).
For gene editing experiments ((C), (D), and (G–J)), n = 2 (Ala146Lys iPSC-RPE and Asn296His iPSC-RPE) and n = 5 (Arg218Cys iPSC-RPE). For electrophysiology experiments ((E), (F), and (K–M)), for +calcium, n = 6 cells for AAVS1, 11 cells for Ala146Lys, 9 cells for Asn296His, 10 cells for Arg218Cys; for no calcium, n = 9 cells for AAVS1, 10 cells for Ala146Lys, 9 cells for Asn296His, 7 cells for Arg218Cys iPSC-RPE (data combined from two replicates). Error bars in (C), (G), and (I) represent mean ± SD; ns = p ≥ 0.05, ∗∗∗ for p < 0.001. Error bars in (F) and (L) represent mean ± SEM; ns = p ≥ 0.05, ∗ for p < 0.05, ∗∗ for p < 0.01.