Significance
CV-A10 is one of the major causative agents of hand-foot-mouth disease, an acute febrile disease occurring mainly in children. To date, there are no effective vaccines or therapeutic drugs available for CV-A10 infection. Moreover, knowledge of the mechanisms of CV-A10 infection and pathogenesis, which are crucial for prevention and treatment of the related diseases, is incomplete. In this work, we provide comprehensive mechanistic insight into CV-A10 infection using the receptor KRM1, demonstrating that KRM1 acts as a two-in-one attachment and uncoating receptor for CV-A10 entry, which mediates both viral attachment at the cell surface and uncoating in the endosome. This work expands our understanding on the mechanism of nonenveloped virus infection and reveals new clues for antiviral intervention.
Keywords: KRM1, Coxsackievirus A10, attachment/uncoating receptor, viral entry, enterovirus
Abstract
KREMEN1 (KRM1) has been identified as a functional receptor for Coxsackievirus A10 (CV-A10), a causative agent of hand-foot-and-mouth disease (HFMD), which poses a great threat to infants globally. However, the underlying mechanisms for the viral entry process are not well understood. Here we determined the atomic structures of different forms of CV-A10 viral particles and its complex with KRM1 in both neutral and acidic conditions. These structures reveal that KRM1 selectively binds to the mature viral particle above the canyon of the viral protein 1 (VP1) subunit and contacts across two adjacent asymmetry units. The key residues for receptor binding are conserved among most KRM1-dependent enteroviruses, suggesting a uniform mechanism for receptor binding. Moreover, the binding of KRM1 induces the release of pocket factor, a process accelerated under acidic conditions. Further biochemical studies confirmed that receptor binding at acidic pH enabled CV-A10 virion uncoating in vitro. Taken together, these findings provide high-resolution snapshots of CV-A10 entry and identify KRM1 as a two-in-one receptor for enterovirus infection.
Enteroviruses are a group of nonenveloped positive-sense RNA viruses belonging to the Enterovirus genus within the family Picornaviridae. Coxsackievirus A10 (CV-A10) is a member of the type-A Enterovirus (EV-A) subgroup. CV-A10 infection is frequently reported to cause hand-foot-mouth disease (HFMD), an acute febrile disease characterized by fever and vesicular exanthema, mostly in the hands, feet, and oral mucosa, posing a tremendous threat to the health of young children globally (1). In addition to HFMD, CV-A10 infection can cause a wide range of clinical manifestations, such as herpangina, aseptic meningitis, and viral meningitis (2). Attention should be paid to emerging CV-A10–associated HFMD and its delayed symptoms to prevent potential outbreaks and improve medical care.
Most enteroviruses enter host cells through a two-receptor mechanism, with the first receptor enabling virion attachment at the cell surface and a second uncoating receptor triggering conformational changes of the viral particle and facilitating genome release into the cytosol (3, 4). However, some enteroviruses, such as poliovirus, use only one receptor to accomplish both steps for cell entry (5). Kringle-containing transmembrane protein 1 (KRM1) was recently identified as an essential entry receptor for CV-A10 and a major subset of EV-As (6); however, the exact molecular mechanism of KRM1 for mediating the entry of these viruses remain incompletely understood.
KRM1 is a ubiquitous membrane-anchored protein located at both cell surface and intracellular membranes. The ectodomain of KRM1 consists of three similarly sized structural domains: an N-terminal Kringle (KR) domain, a poorly folded WSC domain in the middle, and a pseudo–Ig-like CUB domain at the C terminus (7). In previous studies, KRM1 has been extensively characterized as a regulatory ligand of the Wnt/β-catenin signaling pathway that interacts with Dickkopf1 (DKK1) and lipoprotein receptor-related protein 6 (LRP6) (8). The structure of the KRM1-DKK1-LRP6 ternary complex shows that DKK1/LRP6 bind to KRM1 at the N-terminal KR and WSC domains without contacting the C-terminal CUB domain (7). It is unclear which domain of KRM1 is responsible for CV-A10 interaction.
A previous study has demonstrated that CV-A10 binds to the ectodomain of KRM1 at the cell surface and subsequently induces endocytosis (6), suggesting a two-step entry process and a probable pH-dependent uncoating mechanism for CV-A10 infection. Structures of CV-A10 viral particles in different states (i.e., mature virion, metastable A-particle intermediate, and empty capsid) have revealed conserved features for typical enteroviruses, wherein a lipid molecule termed the “pocket factor” is accommodated within a hydrophobic pocket underneath the canyon of viral protein 1 (VP1) subunit. This pocket factor has been suggested as an essential component to maintain the stability of the infectious mature viral particles, and the release of pocket factor is a hypothetical trigger to initiate the uncoating process (4, 9). How the uncoating of CV-A10 is triggered and whether KRM1 is involved in this process are unknown, however.
In this work, we present biochemical evidence indicating that mature CV-A10 virions can bind to KRM1 in both neutral and acidic environments. Structural studies identify KRM1 as a two-in-one receptor for CV-A10 entry and suggest the essential roles of low-pH conditions and receptor binding to initiate CV-A10 uncoating. These findings provide substantial insight into the enterovirus infection process and identify new targets for antiviral intervention.
Results
KRM1 Binds to Mature CV-A10 Virions in Both Neutral and Acidic Conditions.
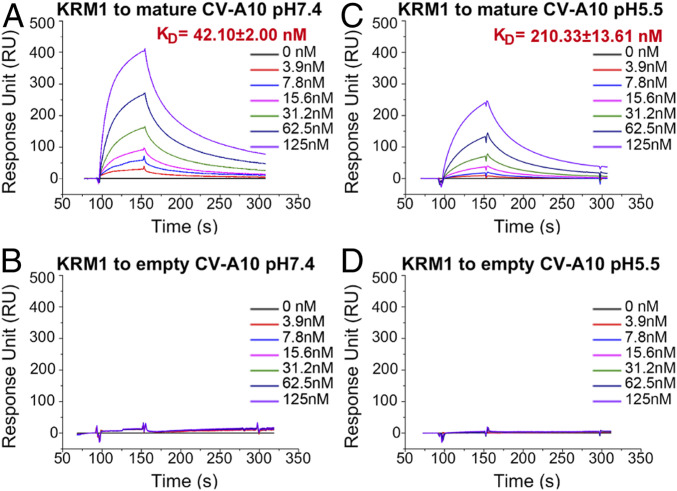
We propagated and purified CV-A10 from human rhabdomyosarcoma (RD) cells. Two bands were observed on a 15 to 45% (wt/vol) sucrose gradient cushion. The top band was composed of empty particles, and the bottom band consisted mainly of mature virions (SI Appendix, Fig. S1 A and B). As shown by surface plasmon resonance (SPR) assays, the ectodomain of KRM1 (residues 23 to 373) selectively bound to mature CV-A10 virions with high affinity (KD = 42 nM) at pH 7.4 (mimicking neutral physiological conditions) (Fig. 1A), whereas the empty particles showed no discernible binding (Fig. 1B). This observation supports the role of KRM1 as an attachment receptor at the cell surface and suggests that empty viral particles may be noninfectious.
Fig. 1.
Binding kinetics between KRM1 and CV-A10 particles in neutral and acidic conditions. (A and C) SPR binding profiles of KRM1 to mature CV-A10 virions at pH 7.4 (A) and pH 5.5 (C). The viral particles were immobilized on the chip surface, and KRM1 was applied as the analyte via serial dilutions. (B and D) Binding profiles of KRM1 to empty CV-A10 capsids at pH 7.4 (B) and pH 5.5 (D). KD values are shown as the mean ± SEM of three independent experiments.
Because many enteroviruses enter host cells through the endosomal pathway and uncoat under acidic conditions, we also tested the binding between KRM1 and mature CV-A10 particles at pH 5.5. Despite displaying a slightly lower affinity (KD = 210 nM) compared with that under neutral conditions, KRM1 could also efficiently bind to mature CV-A10 virions in an acidic environment, suggesting that it may also serve as an uncoating receptor in the endosome (Fig. 1 C and D).
Structures of CV-A10 Viral Particles and Its Complex with KRM1.
To understand how KRM1 mediates CV-A10 entry, we determined the structures of three different types of free CV-A10 viral particles and the complex structures of a mature virion bound to the KRM1 receptor at both neutral and acidic pH. All these structures were determined at atomic or near-atomic resolution, which allowed us to dissect the detailed molecular events during virion–receptor interactions (SI Appendix, Figs. S2–S4).
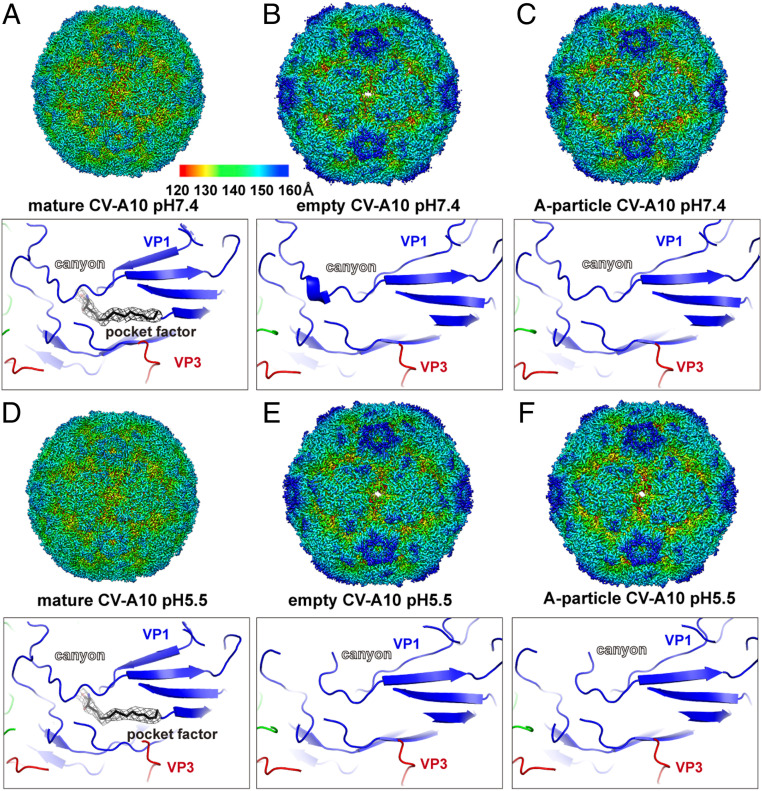
Similar to other enteroviruses, the CV-A10 preparation is composed mainly of mature viral particles (>80%) that are stable under both neutral and acidic conditions (Fig. 2 A and D). In addition, small fractions of A-particles (∼3%) and empty viral capsids (∼10%) were also observed, for which a clear open channel was formed at the two-fold axis (SI Appendix, Fig. S2). Within the mature viral particle, a lipid molecule was accommodated in a pocket beneath the canyon of the VP1 subunit, a signature “pocket factor” of many enteroviruses critical for virion stability (Fig. 2 A and D) (1, 9, 10). Compared with mature viral particles, the empty capsids and A-particle intermediates display conspicuously expanded capsid radii, and the VP4 subunit is dissociated from the capsid layer (Fig. 2). Furthermore, the pocket within VP1 is collapsed due to conformational changes around the canyon, a result of pocket factor ejection to facilitate uncoating (Fig. 2). These observations are consistent with a probable universal mechanism for enterovirus uncoating as observed for poliovirus, Echo viruses, and other related enteroviruses, although the triggering mechanisms for uncoating vary among different viruses (3, 4).
Fig. 2.
Cryo-EM structures of three kinds of CV-A10 particles at pH 7.4 and pH 5.5. (A) Density map of a mature CV-A10 particle at pH 7.4 (3.1 Å) (Upper) and a close-up view of the pocket region within the canyon (Lower). The density map is colored by radius, as indicated by the legend bar. The atomic model is shown in ribbons and colored by chains (VP1, blue; VP2, green; VP3, red). The “pocket factor” is shown as sticks, and the corresponding density map (mesh) is shown at 2.5σ contour. (B–F) Density maps and zoom-in views of atomic models of CV-A10 particles at different states/conditions, shown in the same style as in A.
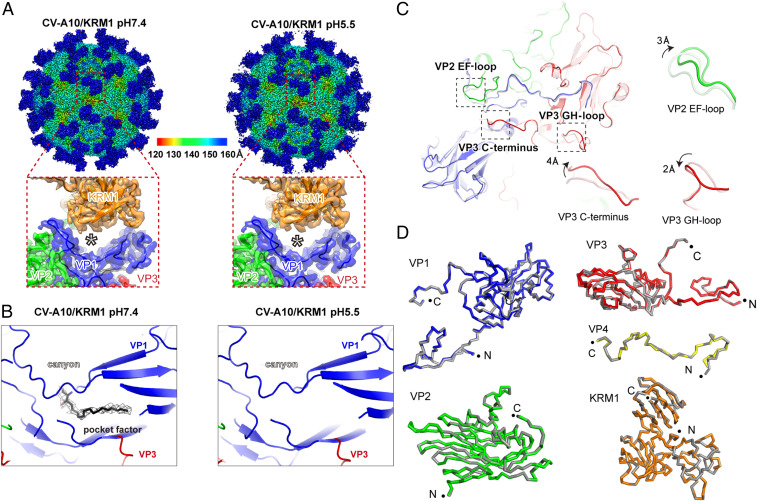
The structures of free CV-A10 particles in different states (mature virions, A-particle intermediates, and empty capsids) at pH 5.5 are highly similar to those of their counterparts at pH 7.4, indicating that free CV-A10 viral particles themselves are stable under different pH conditions (Fig. 2). In the structure of KRM1 bound to CV-A10 at pH 7.4, the lipid was well accommodated, albeit with slightly weaker density compared with that in the free virion, suggesting reduced occupancy due to pocket factor release in some particles (Fig. 3B). Interestingly, the lipid molecules in KRM1-bound viral particles at pH 5.5 became almost invisible in the density map, implying a more profound pocket factor release under acidic conditions (SI Appendix, Fig. S4). These results support the role of KRM1 in mediating CV-A10 attachment at the cell surface and imply that it may also serve as a low-efficiency uncoating receptor at neutral pH, which would become more efficient in the acidic environment. Thus, KRM1 might be a two-in-one receptor for CV-A10 viral entry, responsible for both virion attachment and uncoating.
Fig. 3.
KRM1-bound CV-A10 structures under neutral and acidic conditions. (A) Density maps of the mature CV-A10/KRM1 complex at pH 7.4 (3.0 Å; Left) and pH 5.5 (3.0 Å; Right) viewed along the icosahedral two-fold axes. (Insets) Close-up views of asymmetric units at the receptor-binding interface. (B) Close-up view of the hydrophobic pocket and the “pocket factor” inside, corresponding to pH 7.4 and pH 5.5, respectively. The density map is shown at 2.5σ contour. (C) Atomic models of an asymmetric unit of a CV-A10 virion with (dark colors) or without (light colors) KRM1 binding are aligned. The VP2 EF-loop, VP3 C terminus, and GH-loop are highlighted by dashed boxes, and their close-up views are shown at the side. KRM1 is not shown in the superimposition for clarity. (D) Comparison of each viral subunit in the CV-A10/KRM1 complex at pH 7.4 and pH 5.5. The structures at neutral (gray) and acidic (colored) pH conditions are shown as ribbons and overlaid for each subunit. The N and C termini of each protein are indicated by black dots.
Structural Adaptation for KRM1 Interaction.
In general, the structure of the KRM1-bound viral particle highly resembles the free mature virion; however, several loops at the binding interface display some moderate local conformational changes on receptor engagement (Fig. 3C). The EF-loop in the VP2 subunit, as well as the C terminus and GH-loop of the VP3 subunit, are pulled upward from the viral surface to enable close contact with the receptor (SI Appendix, Figs. S5 and S6). Of note, a long loop (termed the gating loop) within the canyon of the VP1 subunit (around residues 210 to 230) revealed some discernible adjustments in response to KRM1 binding, especially at the bottom region near the lipid exit. The elevation of the VP2 EF-loop deforms its contacts with the VP1 gating loop, which may increase the flexibility of the gating loop around the lipid exit and thereby facilitate release of the pocket factor (SI Appendix, Fig. S6C). This local movement becomes even more significant in the structure of the KRM1-bound virion at pH 5.5 and results in a larger tunnel for pocket factor ejection (SI Appendix, Fig. S6D). These observations may provide an explanation for the structural basis of KRM1-induced pocket factor release of CV-A10 and its greater efficiency in low-pH conditions. In addition, this gating loop becomes highly disordered in the A-particle intermediate and the empty capsid, impairing an important element for KRM1 interaction (SI Appendix, Fig. S6 A and B). Therefore, the KRM1 receptor selectively binds to the mature viral particles but not to the empty capsids or A-particle intermediates (Fig. 1).
Interactions between the CV-A10 Viral Particle and KRM1.
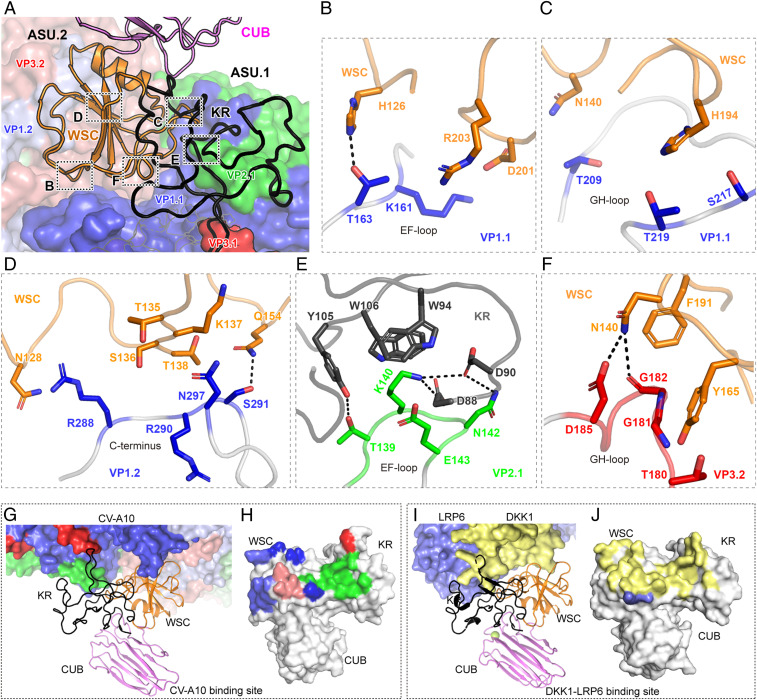
KRM1 stands above the canyon and binds across two adjacent asymmetric units (ASUs) through its KR and WSC domains (Fig. 4A). Two copies of neighboring VP1s are involved in the interaction with KRM1 via different regions. In the VP1 of the first ASU (VP1.1), the EF-loop (residues K161 and T163) and the GH gating loop (residues T209, S217, and T219) form two binding patches with KRM1 on both sides of the canyon (Fig. 4 B and C). Meanwhile, the C terminus of VP1.2 further stabilizes the interaction by contacting the WSC domain of KRM1 (Fig. 4D). In addition, the KR domain of KRM1 covers the VP2.1 EF-loop protrusion (residues 138 to 143) and contributes the majority of strong charged interactions (Fig. 4E). Specifically, residues D88, D90, and Y105 in the KR domain hydrogen-bond to the amidogen of K140 and N142 and to the carbonyl of T139, respectively. The side chains of W94 and W106 in the KR domain contact K140 of VP2.1 through π-cation interactions. Moreover, N140 and Y165 form additional hydrogen bond and hydrophobic interactions with VP3.2 (Fig. 4F and SI Appendix, Table S2). Of note, K140 in VP2.1 and residues 180 to 182 (TGG) in VP3.2 (the key residues participating in receptor binding) are conserved among all KRM1-dependent EV-As identified so far (SI Appendix, Fig. S7), indicating a potentially universal mechanism for receptor binding by these related viruses.
Fig. 4.
Molecular interaction between CV-A10 and KRM1. (A) The overall contacting interface between KRM1 and the CV-A10 virion. Two ASUs of viral proteins are shown in surface models (asu.2 in light colors). VP1, VP2, and VP3 of CV-A10 are shown in blue, green, and red, respectively. KRM1 includes three domains—KR (black), WSC (orange), and CUB (violet)—and is represented as ribbons. (B–F) Interactions between the CV-A10 virus and KRM1 receptor. The proteins are colored by chains with the same color code as in A. The key interacting residues are displayed as sticks and colored by elements. Hydrogen bonds are represented by dashed lines. (G and I) Comparison of the binding mode between CV-A10/KRM1 and the DKK1-LRP6/KRM1 complex. CV-A10, DKK1, and LRP6 are drawn in surface representation. (H and J) KRM1 from the CV-A10/KRM1 complex and the DKK1-LRP6/KRM1 complex is shown in surface representation and colored in gray. The CV-A10, DKK1, and LRP6 binding interfaces are highlighted with the same colors as the interacting protein (VP1.1, blue; VP2.1, green; VP3.1, red; VP1.2, light blue; VP3.2, salmon; DKK1, pale yellow; and LRP6, slate).
To verify our structural analysis, we performed mutagenesis studies of KRM1 by individually replacing the key interacting residues (D90, W106, and Y165) with alanine. All mutants were purified as well-behaved proteins by size-exclusion chromatography (SI Appendix, Fig. S1 D–F); however, their binding affinities to CV-A10 were significantly reduced compared with the wild-type (WT) KRM1 (SI Appendix, Fig. S1 G–J), confirming their critical roles in interactions with the virion.
In addition, KRM1 has also been characterized as a signal transduction receptor on the cell surface that recognizes DKK1 and cooperates with LRP6, forming a ternary complex, to attenuate Wnt/β-catenin signaling activation (7, 8). Compared with the CV-A10/KRM1 complex, we found that KRM1 uses a highly similar interface to bind CV-A10 or DKK1/LRP6, suggesting that these interactions are mutually exclusive (Fig. 4 G–J and SI Appendix, Fig. S8).
KRM1-Mediated CV-A10 Uncoating at Acidic pH In Vitro.
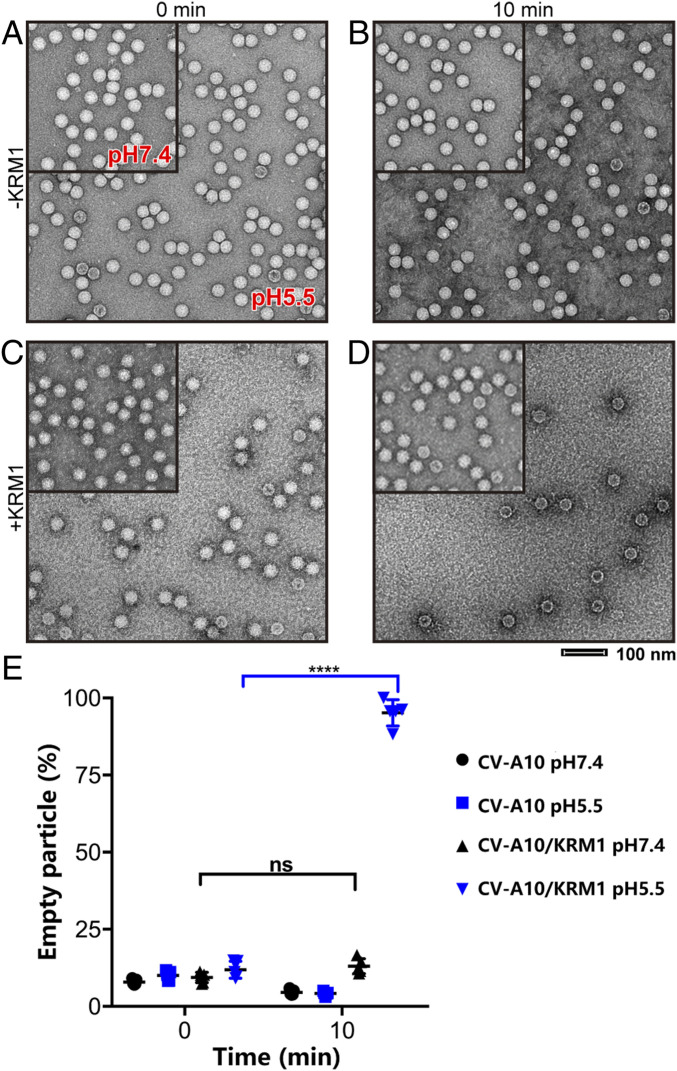
Because the acidic pH conditions enable efficient pocket factor release of CV-A10 on KRM1 interaction, we sought to investigate whether soluble KRM1 could directly induce CV-A10 virion uncoating in vitro. To do so, we incubated mature CV-A10 particles with soluble KRM1 in both neutral (pH 7.4) and acidic (pH 5.5) conditions at physiological temperature (37 °C) and compared the virion morphologies by negative stain electron microscopy (EM). The mature CV-A10 particles alone did not exhibit significant changes under these treatments, consistent with the cryo-EM structures (Fig. 2). In the presence of KRM1, a substantial proportion of viral particles transformed into empty capsids under acidic pH conditions, whereas no discernible changes occurred at pH 7.4 (Fig. 5).
Fig. 5.
KRM1-dependent uncoating of CV-A10 in vitro. (A–D) Representative negative stain EM images of four CV-A10 virus groups without (A and B) or with (C and D) KRM1 proteins and exposed to neutral pH (pH 7.4, Upper Left) or acidic pH (pH 5.5) treatments. Samples were negatively stained immediately after mixing or after a 10-min incubation at 37 °C. (E) The percentage of empty particles for each group is displayed in the interleaved scatterplot, with a horizontal line indicating the median. The significance of differences was tested by Tukey’s multiple comparison test. ****P < 0.0001; ns, not significant.
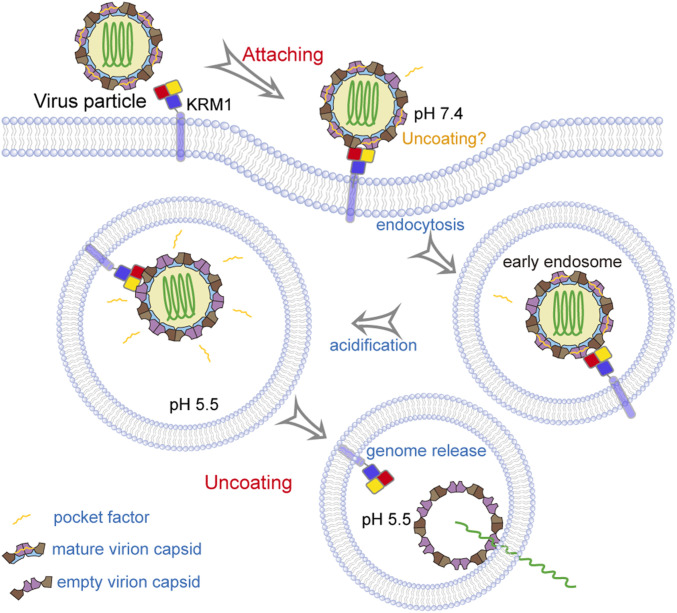
These observations demonstrate that KRM1 is a two-in-one receptor for CV-A10 entry that can mediate both viral attachment and uncoating to enable virus entry. Interestingly, the low-pH condition is much more favorable than neutral conditions for viral uncoating, suggesting that the viruses may infect cells mainly through the endosomal pathway. As reported previously, the binding of KRM1 to CV-A10 would trigger the endocytosis of viral particles into the endosome (6), which would then facilitate the uncoating process in the acidic environment. In addition, as the binding of KRM1 to CV-A10 at neutral conditions resulted in low-efficiency release of the pocket factor, the uncoating process may also occur at the cell membrane as a minor alternative pathway for cell entry (Fig. 6).
Fig. 6.
KRM1 is a two-in-one receptor for CV-A10 entry into the cell. KRM1 is anchored on the cell membrane, and the three structural domains (KR, WSC, and CUB) are displayed in red, yellow, and slate, respectively. The four structural proteins (VP1, VP2, VP3, and VP4) within the viral capsid are indicated in violet purple, brown, chocolate, and sky blue, respectively. The viral RNA genome and the “pocket factor” are shown in forest green and orange, respectively.
Discussion
Enteroviruses infect host cells by recognizing membrane receptors, followed by uncoating at either the cell surface or in the endosome (11). Some of these viruses bind to different receptors for attachment and uncoating, whereas several viruses use a single receptor for both steps. Many EV-Bs use CD55 for attachment to the cell surface and switch to the endosomal receptor FcRn to trigger uncoating in acidic conditions (3, 12). A special example among the EV-Cs is poliovirus, which uses CD155 (PVR) as a bifunctional receptor for both attachment and uncoating (5). As for EV-As, the scavenger receptor class B member 2 (SCARB2) has been identified as the receptor for EV-A71 and CV-A16 (13). A recent structural study also revealed an unexpected binding mode between EV-A71 and SCARB2, where SCARB2 binds to the “southern rim” of the canyon, interacting with VP1 and VP2 to initiate uncoating (SI Appendix, Figs. S8B and S10). Sequence alignment for SCARB2-dependent EV-As shows low conservation in the SCARB2-binding residues (14), suggesting remarkable variability for interactions with the same receptor. In this study, we found that KRM1 binds across two ASUs and interacts with all viral proteins in the outer shell, for which the binding interface is moderately conserved among all KRM1-dependent EV-As characterized so far (SI Appendix, Fig. S7). These findings suggest that KRM1 displays a more uniform recognition pattern by different EV-As.
As observed in many enteroviruses, uncoating receptors usually bind to the canyon of the virion via their Ig-like domain and trigger pocket factor release to initiate conformational changes for uncoating (3, 11). However, KRM1 binds to CV-A10 above the canyon through its KR and WSC domains instead of through the CUB pseudo Ig-like domain (SI Appendix, Fig. S9A). It covers the canyon on both sides but does not penetrate inside to directly interact with the canyon, which is unique compared with the more prevalent canyon-penetrating receptors. It is interesting that KRM1 interacts with the distal region of the gating loop to potentially alter its conformation around the lipid exit within the canyon, thereby facilitating release of the “pocket factor.” How the acidic environment triggers more significant conformational changes of the gating loop to accelerate pocket factor release remains unclear. A possible mechanism is that the low pH condition leads to protonation of histidine residues around the canyon, which may perturb the interaction networks of the gating loop to its anchor partners and increase its flexibility. The role of histidine residues serving as pH sensors is well established for the fusogenic glycoproteins of enveloped viruses (15), and it may also apply to the pH-dependent entry processes of nonenveloped viruses. In addition, temperature may affect the stability of virion–receptor interactions, further modulating the uncoating efficiency. In the cryo-EM images, the relative abundance of empty capsids and A-particles did not exhibit obvious changes in the presence of KRM1, irrespective of the pH condition (SI Appendix, Fig. S2). This observation suggests that KRM1 binding may temporarily stabilize the virion structure after pocket factor release at low temperature (4 °C in this case), thus preventing conformational changes of the viral capsid to initiate genome release. In contrast, the receptor would be prone to dissociate at physiological temperature from the metastable intermediate, resulting in efficient genome release from the virions (Fig. 5).
During the preparation of this manuscript, Zhao et al. (16) reported a similar near-atomic resolution (3.9 Å) structure of KRM1 bound to CV-A10 at pH 8 and suggested that KRM1 could potentially induce pocket factor release of the virion. This is consistent with our observation that KRM1 binding slightly reduced the occupancy of pocket factor in the density map at pH 7.4 compared with the free virion. However, the authors did not investigate the effects of different pH conditions on virion–receptor interactions and thus could not comprehensively define the mechanism of virus entry. Moreover, our structures and biochemical evidence allowed better resolution of the mechanistic details at different stages of virus entry and provided more insight into the role of KRM1 for mediating entry by CV-A10 and related enteroviruses (Fig. 6).
It has been shown that viruses tend to use the cell signaling and regulatory pathways to induce endocytosis and facilitate subsequent invasion (17, 18). On the other hand, KRM1 is also involved in the conserved Wnt/β-catenin pathway, in which it interacts with DKK1 and LRP6 to form a ternary complex and induce endocytosis. Previous studies have shown that KRM1 binds to DKK1-LRP6 via its KR and WSC domains, and the introduction of N-linked glycans on D90 also decreases the binding capacity of KRM1 to DKK1 (7). The key interacting residues involved in the CV-A10–KRM1 interaction significantly overlap with the interface for DKK1/LRP6 binding. Thus, CV-A10 could possibly mimic DKK1-LRP6 for KRM1 binding and internalization to enable cell entry.
In addition, a previously reported CV-A10/Fab complex structure revealed that the VP1 C terminus, VP2 EF-loop, and VP3 AB-loop constitute candidate neutralization epitopes (1, 19) (SI Appendix, Fig. S9C). Here we found that the VP2 EF-loop (residues 138 to 143) contributed the majority of the strong charged interactions with KRM1, suggesting that this region might be a hotspot of interference with the receptor-binding process to block virus entry. In addition, the KRM1-binding interface on CV-A10 covers a broad region with multiple contacting patches, which may imply potent epitopes for developing vaccines and therapeutic antibodies.
In summary, here we present the molecular basis for KRM1-mediated CV-A10 entry into host cells and propose that KRM1 is a two-in-one attachment and uncoating receptor. Attachment at neutral pH enables the virus association with cells at the plasma membrane, and the low-pH conditions in the endosome accelerate pocket factor release to initiate the uncoating process. These findings provide systematic mechanistic insight into the entry of CV-A10 and related viruses, which may constitute an important basis for developing antiviral therapeutics.
Materials and Methods
Virus Production and Purification.
The CV-A10 strain HB09-035 was first isolated in 2009 in Hebei Province, China. Human RD cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 100 U/mL penicillin-streptomycin. RD cells were grown to 95% confluence and infected with CV-A10 at a multiplicity of infection (MOI) of 0.1. Infected cells were incubated at 37 °C for 36 h, at which point ∼90% of cells displayed a cytopathic effect. The cultures were harvested and centrifuged at 12,000 × g for 60 min to remove cell debris.
Viruses from the supernatant were purified as described previously (3). In brief, the virus was precipitated with a 30% (wt/vol) sucrose cushion and resuspended in phosphate-buffered saline (PBS). Two bands were observed on the 15 to 45% (wt/vol) sucrose density gradient. The sample was fractionized into aliquots of 200 μL from the top to the bottom of the sucrose cushion. Each fraction was quantified by measuring the optical density (OD) at 260 and 280 nm with a NanoDrop spectrophotometer and further visualized by negative stain EM and sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The top band was composed mainly of empty capsid particles (OD260/OD280 = 0.9), and the bottom band consisted mainly of mature virions (OD260/OD280 = 1.7) (SI Appendix, Fig. S1 A and B). Then the two bands were individually dialyzed against 1×PBS (pH 7.4) buffer.
Protein Production and Purification.
The construct of soluble human KRM1 protein has been described previously (7). In brief, the human KRM1 ectodomain (A23-G373) was synthesized and cloned into pCMV3 (Sino Biological) with a human IL-2 signal sequence at the N terminus and 10 histidines at the C terminus using the HindIII and BamHI sites, respectively. The recombinant plasmid was transiently transfected into HEK293T cells for KRM1 expression. The supernatant was harvested at 7 d posttransfection and centrifuged to remove cell debris, and the protein was subsequently purified by Ni-nitrilotriacetic acid chromatography (GE Healthcare Life Sciences). The target proteins were eluted using 300 mM imidazole dissolved in PBS. The fractions were then dialyzed into PBS, followed by size-exclusion chromatography using a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences). The human KRM1 mutant plasmids were constructed by site-directed mutagenesis, for which the identified key residues involved in CV-A10 binding (D90A, W106A, and Y165A) were mutated separately, and the proteins were purified by the same methods (SI Appendix, Fig. S1 C–F).
SPR Assay.
The binding kinetics and affinity of KRM1 to CV-A10 were analyzed by SPR using a BIAcore 3000 biosensor (GE Healthcare Life Sciences) at room temperature (25 °C). All proteins used for kinetic analyses were exchanged into PBST buffer through gel filtration. Mature CV-A10 virus was biotinylated and immobilized on a SA chip to approximately 6,000 response units. The soluble ectodomain of KRM1 and the D90A, W106A, Y165A mutant proteins were serially diluted with PBST, sequentially injected into the chip at a flow rate of 30 μL/min for 1 min, and then allowed to dissociate over 2 min. The binding curve at zero concentration of protein was subtracted as a blank from each experimental curve. After each cycle, a short injection of 3 M MgCl2 was used to regenerate the sensor surface. Data were analyzed, and kinetic constants were estimated using BIAevaluation 4.1 software (GE Healthcare Life Sciences).
Cryo-EM Sample Preparation and Data Collection.
Purified mature CV-A10 virus was incubated with excess KRM1 at final concentrations of 2 mg/mL and 0.5 mg/mL for 10 min at 4 °C. The CV-A10 viral particle or CV-A10/KRM1 complex samples (3 μL), at pH 7.4, were placed on a freshly plasma-cleaned lacey carbon grid and allowed to adsorb to the grid for 60 s, followed by blotting with a filter paper for 3 s before plunge-freezing using a Vitrobot Mark IV (FEI) operated at 4 °C and 100% humidity. For the acidic condition samples, CV-A10/KRM1 samples were incubated in low pH buffer [20 mM 2-(N-morpholino) ethanesulphonic acid and 150 mM NaCl, pH 5.5] for 10 min, then applied onto the grid as described above.
Cryo-EM data were collected using either a 300-kV FEI Titan Krios (for CV-A10 alone) or a 200-kV FEI Talos Arctica (for CV-A10/KRM1 complex) electron microscope equipped with a Gatan K2 Summit direct electron detector. Images were recorded in superresolution counting mode with a calibrated pixel size of 1.35 Å /1.32 Å. Defocus values varied from -1.5 to −2.5 μm. Each image was dose-fractionated to 32 frames with a total electron dose of 40 e−/Å2.
Image Processing.
The beam-induced image drift and anisotropic magnification of each image stack were corrected with the MotionCor2 program (20). Initial parameters of the contrast transfer function (CTF) were determined by CTFFIND4 (21) at the micrograph level. Particles were automatically picked with Gautomatch (by K. Zhang). The particles were extracted as 4× binned particles and subjected to reference-free 2D classification with Relion 3.0 (22). Ice contamination, damaged particles, and low-contrast images were removed after multiple rounds of classification and subset selection. The clean dataset was recentered and reextracted for subsequent 3D classification with the density map of free CV-A10 mature virions (EMD-9674) as the reference model, which was low-pass filtered to 60 Å before image alignment. The angular assignment was initiated with a global search and proceeded with a restricted local search based on the estimation of alignment accuracy. For each dataset, viral particles in different states were observed with clear features of secondary structural elements. The most distinguished classes with the best alignment accuracy were selected for 3D refinement for each structure. After the initial refinement with 2× binned particles, the full-size particle images were further recentered and reextracted for a second round of 3D refinement. At the final stage, CTF refinement was performed with Relion 3.0 (22) to counteract the defocus variation of each particle, and the dose-weighted images were calculated with MotionCor2 (20) to balance the effects of radiation damage. An additional round of 3D refinement was performed with per-particle CTF values and dose-weighted images to calculate the final reconstruction. The numbers of initial and final particles and the composition of different conformational particles in each dataset are shown in SI Appendix, Table S1 and Fig. S2. Local resolution was estimated using ResMap (23) for each reconstruction.
Model Building and Refinement.
The crystal structure of KRM1 (Protein Data Bank [PDB] ID code 5FWS) and the cryo-EM structure of CV-A10 (PDB ID code 6ACU) were used as the initial model to fit the CV-A10/KRM1 density map (pH 7.4, 3.0 Å; pH 5.5, 3.0 Å) using Chimera (24). After initial fitting, the model was manually adjusted in Coot (25) to improve map fitting and update the sequence registers. The density map of an ASU was then segmented using Chimera (24) and placed into a pseudo-crystallographic unit cell (P1 space group) for model refinement. The atomic model was refined against the ASU map in real space using Phenix (26), with secondary structure restraints applied. Model statistics (including bond lengths, bond angles, all-atom clash, and rotamer outliers), and Ramachandran plot statistics were monitored during the refinement procedure. After several rounds of iterative model building and refinement, the model coordinates converged and fit well in the density map by visual inspection. At this point, the individual B factors and occupancies of each atom were refined using the standard reciprocal space refinement procedure in Phenix (26). The stereochemical quality of the final model was assessed using MolProbity (27). The statistics for 3D reconstruction and model refinement are summarized in SI Appendix, Table S1.
Structure Analysis and Visualization.
The reconstructed maps and atomic models were visualized using Chimera (24) and analyzed using the wrapped applications. All EM density figures were rendered with Chimera, and atomic model representations were generated with the PyMOL molecular graphics system (https://pymol.org/2/).
In Vitro Uncoating Assays and Negative Stain EM.
Purified mature CV-A10 virus was incubated alone or mixed with KRM1 protein at a molar ratio of 1:70 in pH 7.4 or pH 5.5 buffers. Samples were negatively stained immediately after mixing or after a 10-min incubation at 37 °C. Grids were examined with an FEI Tecnai G20 transmission electron microscope operated at an accelerating voltage of 120 kV. Images were recorded at a magnification of 29,000×, resulting in a pixel size of 3.76 Å/pixel. Defocus values varied from −2 to −4 μm. Each image was exposed with a total dose of 60 e−/Å2 and recorded with a BM-Eagle CCD camera. The empty particles were quantified by visual inspection. The statistics for each sample were calculated with GraphPad Prism using approximately 2,000 total particles from randomly selected images. The significance of difference was tested by Tukey’s multiple comparison implemented in GraphPad Prism.
Supplementary Material
Acknowledgments
We thank Dr. Yong Zhang (Chinese Center for Disease Control and Prevention) who kindly provided the CV-A10 HB09-035 strain (GenBank accession no. MT263729). We thank all staff members in the Center of Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences (CAS), Beijing, and the Cryo-EM Center of Southern University of Science and Technology for assistance with cryo-EM data collection. We are grateful to the staff members in the EM department of the State Key Laboratory of Membrane Biology, Institute of Zoology, CAS, Beijing, for technical support in electron microscope operation. Special thanks to Drs. Zheng Fan and Wei Zhang of the Institute of Microbiology, CAS for assisting with the SPR experiments. This work was supported by the National Science and Technology Major Projects (2018ZX09711003), the Strategic Priority Research Program of the CAS (XDB29010202), the National Science and Technology Major Project (2018ZX10733403), the Technological Innovation Project of Shanxi Transformation and Comprehensive Reform Demonstration Zone (2017KJCX01), the Beijing Nova Program of Science and Technology (Z191100001119030), and the Youth Innovation Promotion Association of the CAS (20200920). R.P. is supported by the Young Elite Scientist Sponsorship Program of the China Association for Science and Technology (2018QNRC001).
Footnotes
The authors declare no competing interest.
Data deposition: The density maps have been deposited to the Electron Microscopy Data Bank with accession nos. EMD-30253 (mature CV-A10, pH 7.4), EMD-30254 (mature CV-A10, pH 5.5), EMD-30287 (A-particle CV-A10, pH 7.4), EMD-30290 (A-particle CV-A10, pH 5.5), EMD-30291 (empty CV-A10, pH 7.4), EMD-30292 (empty CV-A10, pH 5.5), EMD-30259 (CV-A10/KRM1, pH 7.4), and EMD-30260 (CV-A10/KRM1, pH 5.5). The coordinates of the corresponding atomic models have been deposited to the Protein Data Bank with ID codes 7BZN (mature CV-A10, pH 7.4), 7BZO (mature CV-A10, pH 5.5), 7C4T (A-particle CV-A10, pH 7.4), 7C4W (A-particle CV-A10, pH 5.5), 7C4Y (empty CV-A10, pH 7.4), 7C4Z (empty CV-A10, pH 5.5), 7BZT (CV-A10/KRM1, pH 7.4), and 7BZU (CV-A10/KRM1, pH5.5). The sequence of the CV-A10 HB09-035 strain has been deposited in the GenBank database (accession no. MT263729).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005341117/-/DCSupplemental.
Data Availability.
The density maps have been deposited to the Electron Microscopy Data Bank with accession codes EMD-30253 (mature CV-A10, pH 7.4), EMD-30254 (mature CV-A10, pH 5.5), EMD-30287 (A-particle CV-A10, pH 7.4), EMD-30290 (A-particle CV-A10, pH 5.5), EMD-30291 (empty CV-A10, pH 7.4), EMD-30292 (empty CV-A10, pH 5.5), EMD-30259 (CV-A10/KRM1, pH 7.4), and EMD-30260 (CV-A10/KRM1, pH 5.5). The coordinates of the corresponding atomic models were deposited to the Protein Data Bank with ID codes 7BZN (mature CV-A10, pH 7.4), 7BZO (mature CV-A10, pH 5.5), 7C4T (A-particle CV-A10, pH 7.4), 7C4W (A-particle CV-A10, pH 5.5), 7C4Y (empty CV-A10, pH 7.4), 7C4Z (empty CV-A10, pH 5.5), 7BZT (CV-A10/KRM1, pH 7.4), and 7BZU (CV-A10/KRM1, pH 5.5).
References
- 1.Zhu R. et al., Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10. Sci. Adv. 4, eaat7459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bian L. et al., Hand, foot, and mouth disease associated with coxsackievirus A10: More serious than it seems. Expert Rev. Anti Infect. Ther. 17, 233–242 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Zhao X. et al., Human neonatal Fc receptor is the cellular uncoating receptor for Enterovirus B. Cell 177, 1553–1565.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossmann M. G., He Y., Kuhn R. J., Picornavirus-receptor interactions. Trends Microbiol. 10, 324–331 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Strauss M. et al., Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J. Virol. 89, 4143–4157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staring J. et al., KREMEN1 is a host entry receptor for a major group of enteroviruses. Cell Host Microbe 23, 636–643.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Zebisch M., Jackson V. A., Zhao Y., Jones E. Y., Structure of the dual-mode wnt regulator kremen1 and insight into ternary complex formation with LRP6 and Dickkopf. Structure 24, 1599–1605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao B. et al., Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 417, 664–667 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Chen J. et al., Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses. Cell Discov. 5, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L. et al., Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating. Nat. Commun. 9, 4985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells A. I., Coyne C. B., Enteroviruses: A gut-wrenching game of entry, detection, and evasion. Viruses 11, 460–479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu S. et al., Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn. Chin. Sci. Bull. 65, 67–79 (2020). [Google Scholar]
- 13.Yamayoshi S. et al., Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Zhou D. et al., Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. Nat. Microbiol. 4, 414–419 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Peng R. et al., Structures of human-infecting Thogotovirus fusogens support a common ancestor with insect baculovirus. Proc. Natl. Acad. Sci. U.S.A. 114, E8905–E8912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y. et al., Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10. Nat. Commun. 11, 38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cossart P., Helenius A., Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 6, a016972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain K. M., Leong K. L., Ng M. M., Chu J. J., The essential role of clathrin-mediated endocytosis in the infectious entry of human enterovirus 71. J. Biol. Chem. 286, 309–321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai W. et al., Recombinant virus-like particle presenting a newly identified coxsackievirus A10 neutralization epitope induces protective immunity in mice. Antiviral Res. 164, 139–146 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimanius D., Forsberg B. O., Scheres S. H. W., Lindahl E., Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen E. F. et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams P. D. et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen V. B. et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The density maps have been deposited to the Electron Microscopy Data Bank with accession codes EMD-30253 (mature CV-A10, pH 7.4), EMD-30254 (mature CV-A10, pH 5.5), EMD-30287 (A-particle CV-A10, pH 7.4), EMD-30290 (A-particle CV-A10, pH 5.5), EMD-30291 (empty CV-A10, pH 7.4), EMD-30292 (empty CV-A10, pH 5.5), EMD-30259 (CV-A10/KRM1, pH 7.4), and EMD-30260 (CV-A10/KRM1, pH 5.5). The coordinates of the corresponding atomic models were deposited to the Protein Data Bank with ID codes 7BZN (mature CV-A10, pH 7.4), 7BZO (mature CV-A10, pH 5.5), 7C4T (A-particle CV-A10, pH 7.4), 7C4W (A-particle CV-A10, pH 5.5), 7C4Y (empty CV-A10, pH 7.4), 7C4Z (empty CV-A10, pH 5.5), 7BZT (CV-A10/KRM1, pH 7.4), and 7BZU (CV-A10/KRM1, pH 5.5).