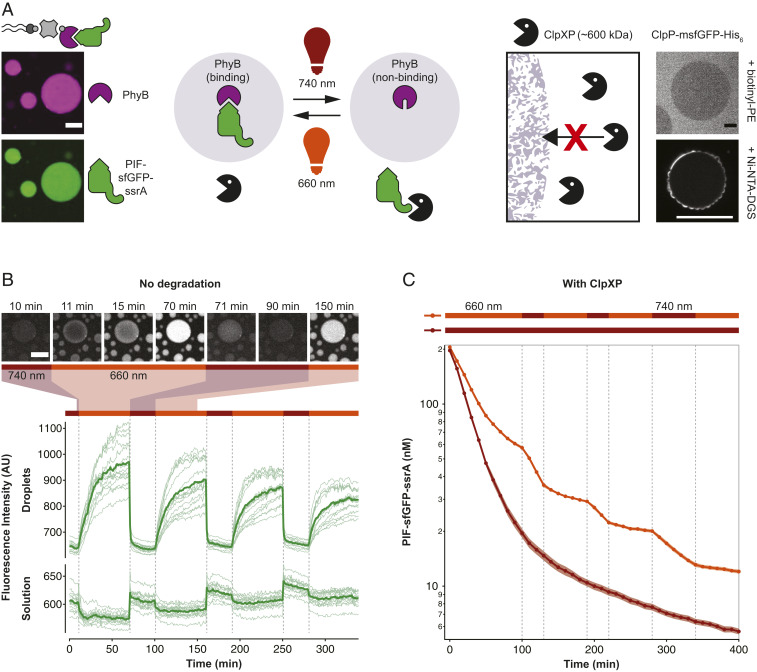
Fig. 5.
Light-dependent protein capture and release to dynamically control degradation rates. (A) Design of the light-controlled protein degradation reaction. (Left) Biotin-streptavidin–mediated binding of light-responsive phytochrome B (PhyB) and capture of the protease substrate PIF-sfGFP-ssrA in 660-nm light. (Right) Exclusion of ClpXP protease from lipid sponge droplets. ClpP-msfGFP-His6 is excluded from droplets doped with biotinyl-PE and is unable to enter the droplets when targeted inside via Ni-NTA-DGS. (B) Light-controlled release and capture of PIF-sfGFP-ssrA by PhyB-loaded droplets. In this experiment, no ClpXP was added. Traces show fluorescence intensities of droplets and solution over time in response to changing illumination conditions. Bars above indicate 660- and 740-nm conditions. Intensities were extracted from a total of 15 droplets in five time-lapse movies (thin lines) and averaged (bold lines). Selected images from a time-lapse movie are shown on Top. (C) Light-controlled switching between protection and degradation of PIF-sfGFP-ssrA by ClpXP. Degradation of PIF-sfGFP-ssrA was monitored by reading the fluorescence of the reaction every 10 min. Reactions were performed in triplicates. Shown is the average (bold) and the SD (shaded areas). (Scale bars, 20 µm.)