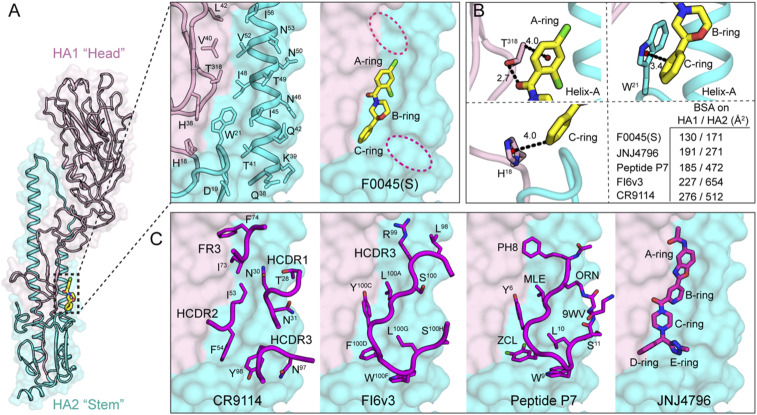
Fig. 4.
Structure and molecular interactions of small-molecule F0045(S) with influenza A HA. (A) Crystal structure of influenza A HA from group 1 H1N1 H1/PR8 (HA1, light pink and HA2, cyan cartoon with molecular surface representation) in complex with small-molecule F0045(S) (carbon, oxygen, nitrogen, and chlorine are represented in yellow, red, blue, and green, respectively). One protomer of the H1/PR8 trimer is represented. F0045(S)-binding site residues on the molecular surface of H1/PR8 HA shown in sticks on a backbone tube representation. Additional hydrophobic pockets around F0045(S) are highlighted in dotted red ovals. (B) Molecular interactions in the F0045(S)-H1/PR8 HA complex. The centroids of the rings are shown in a red sphere, hydrogen bonds are shown in black dotted lines, and distances are measured in angstroms (Å). Buried surface area (BSA) of F0045(S) and other inhibitors on HA (HA1/HA2). (C) Overlay of structures of F0045(S)-H1/PR8 with HA-interacting loop residues from the Fabs of HA-Fab complexes of CR9114 (PDB ID 4FQI), FI6v3 (PDB ID 3ZTN), with cyclic peptide P7 (PDB ID 5W6T), and small-molecule JNJ4796 (PDB ID 6CF7). Antibody CDR loops and cyclic peptide are represented as a backbone tube with side chains as sticks and JNJ4796 in stick representation, respectively.