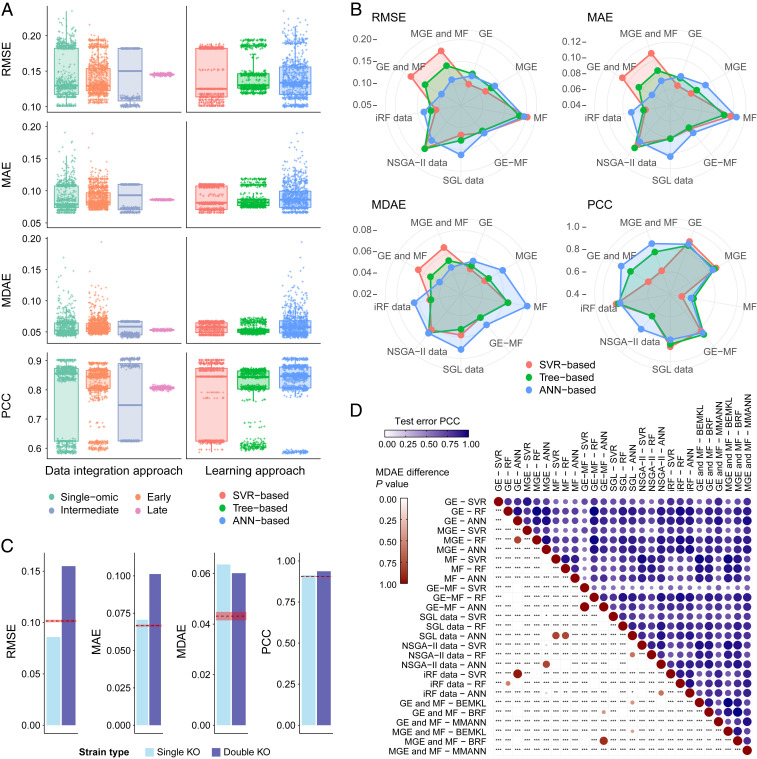
Fig. 3.
Machine-learning yeast growth prediction results. (A) Comparison of model predictive performance across data integration strategy and machine-learning model type. Intermediate integration is overall the most effective approach and notably better than single-omic models. Concomitantly, ANN- and SVR-based techniques appear generally more effective than tree-based techniques. (B) Comparison of model accuracy for all dataset–learning algorithm combinations, corresponding to numeric results shown in Table 1. The MMANN using both GE and MF profiles is overall the most accurate model, followed by GE-based SVR. (C) Error scores on the experimentally independent test set. Dashed red lines represent the corresponding error score on the main test set, while shaded areas represent their associated SD. (D) In blue, Pearson’s correlation between error score vectors on the test set, for each pair of data–method combination. In red, P values are shown of Wilcoxon rank-sum tests assessing the significance of MDAE differences, for each pair of data–method combination. *, **, and *** represent significance at thresholds of 0.05, 0.01, and 0.001, respectively, rescaled by Bonferroni correction.