Significance
Repeat-associated non-AUG (RAN) proteins accumulate in patient brains and contribute to a growing number of neurodegenerative diseases. There is an urgent need to understand why expression of these proteins does not require canonical or near-cognate AUG start codons and to develop ways to block RAN protein production. We show several types of repeat-expansion RNAs activate the double-stranded RNA-dependent protein kinase (PKR) pathway and that blocking PKR reduces RAN protein levels in cells. PKR is activated in C9orf72 ALS/FTD human and mouse brains and PKR inhibition using AAV-PKR-K296R or the FDA-approved drug metformin decreases RAN protein levels and improves disease in ALS/FTD mice. Targeting PKR using gene therapy or metformin are promising therapeutic approaches for C9orf72 ALS/FTD and other expansion diseases.
Keywords: metformin, RAN translation, C9orf72, PKR, ALS/FTD
Abstract
Repeat associated non-AUG (RAN) translation is found in a growing number of microsatellite expansion diseases, but the mechanisms remain unclear. We show that RAN translation is highly regulated by the double-stranded RNA-dependent protein kinase (PKR). In cells, structured CAG, CCUG, CAGG, and G4C2 expansion RNAs activate PKR, which leads to increased levels of multiple RAN proteins. Blocking PKR using PKR-K296R, the TAR RNA binding protein or PKR-KO cells, reduces RAN protein levels. p-PKR is elevated in C9orf72 ALS/FTD human and mouse brains, and inhibiting PKR in C9orf72 BAC transgenic mice using AAV-PKR-K296R or the Food and Drug Administration (FDA)-approved drug metformin, decreases RAN proteins, and improves behavior and pathology. In summary, targeting PKR, including by use of metformin, is a promising therapeutic approach for C9orf72 ALS/FTD and other expansion diseases.
More than 50 different neurological diseases are caused by microsatellite repeat expansions located within 5′ and 3′ untranslated regions (UTRs), introns, and protein-coding regions. Expansion mutations can be bidirectionally transcribed (1, 2), and many of these mutated genes have been shown to produce proteins, which are synthesized from multiple reading frames in the absence of AUG or near-cognate AUG initiation codons through a process called repeat associated non-AUG (RAN) translation (3). RAN translation, initially discovered in spinocerebellar ataxia type 8 (SCA8) and myotonic dystrophy type 1 (DM1), has now been reported in 10 different repeat expansion disorders including C9orf72 amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (3–7). Knowledge of the molecular mechanisms of RAN translation is important for understanding the role of RAN proteins in disease and for the development of therapeutic strategies. Previous studies have shown that RNA structure and flanking sequences affect RAN protein synthesis (3) and, more recently, that RAN protein levels increase under stress conditions, including ER stress in vitro (8, 9). Mutant repeat expansion RNAs (e.g., CAG, CUG, CGG, G4C2, and G2C4) that form hairpin or G-quadruplex structures are a common theme in RAN protein diseases (3, 7, 10, 11). Hairpin forming HIV1 mRNA (12) and CUG expansion RNAs, which form imperfect hairpins, were shown to activate the double-stranded RNA-dependent protein kinase (PKR) (13). PKR is a member of a family of serine-threonine kinases that phosphorylate the translation initiation factor eIF2 on its α subunit (eIF2α) (14). This impairs translational initiation of most proteins, while inducing the translation of a selective set of mRNAs, including transcriptional regulators. Other members of the eIF2α kinase family are as follows: PERK (PKR-like ER kinase), GCN2 (general control non-derepressible-2), and HRI (heme-regulated inhibitor). Here, we show that a variety of structured RAN-positive repeat expansion RNAs activate the PKR pathway and that inhibition of PKR decreases RAN protein levels in cell culture and C9orf72 BAC transgenic ALS/FTD (C9-BAC) mice (15). Additionally, we demonstrate that the widely used Food and Drug Administration (FDA)-approved diabetes drug metformin inhibits PKR phosphorylation and activation, decreases RAN protein levels, and improves phenotypes in C9orf72 ALS/FTD BAC mice.
Results
RAN Translation Highly Regulated by PKR.
Given that repeat expansions undergo RAN translation and CUG repeats activate PKR (13), we investigated whether other RAN positive repeat expansion RNAs similarly activate PKR and the role of PKR in RAN translation. First, we show that in HEK293T cells transfected with repeat expansion constructs (SI Appendix, Fig. S1A) expressing several types of disease relevant repeat expansion RNAs [(CAG)105, (CCUG)128, (CAGG)137, or (G4C2)120] show an increase in the levels of phospho-PKR (p-PKR) at sites indicative of PKR activation (T446 and T451) (SI Appendix, Fig. S1B). Using an in vitro PKR kinase assay (16), we show that p-PKR levels are increased in mouse embryonic fibroblasts (MEFs) harboring a single (C9-500) or two copies (C9-500/32) of the G4C2 expansion (Fig. 1A). Consistent with the C9 MEFs, p-PKR levels are also increased in liver tissue from C9-500 mice (Fig. 1B). Similarly, immunofluorescence staining showed p-PKR levels are higher in the CA region of the hippocampus in a cohort of phenotypic C9-500 vs. NT control mice (Fig. 1C and SI Appendix, Fig. S2). Additionally, and similar to the results in Huntington disease brains (17), we show p-PKR levels are increased in hippocampal tissue from C9 patient autopsy brains (Fig. 1D). While protein blots show a similar trend toward increased phospho-eIF2α (p-eIF2α) in C9-500 mouse brains compared to nontransgenic (NT) mice (SI Appendix, Fig. S3), the results were not significant. Additional cell culture experiments show that PKR activation is dependent on the levels of G4C2 expansion RNAs (SI Appendix, Fig. S4A). Because short CAG small interfering (si) RNAs have been reported in Huntington’s disease (18), we tested but found no evidence that p-PKR levels were increased in HEK293T cells transfected by short si (CAG)7 RNAs (SI Appendix, Fig. S4B). We also found no evidence that p-PKR levels could be elevated by polyAla proteins expressed from a CAG expansion construct with an AUG initiation codon in the polyAla reading frame (SI Appendix, Fig. S4C).
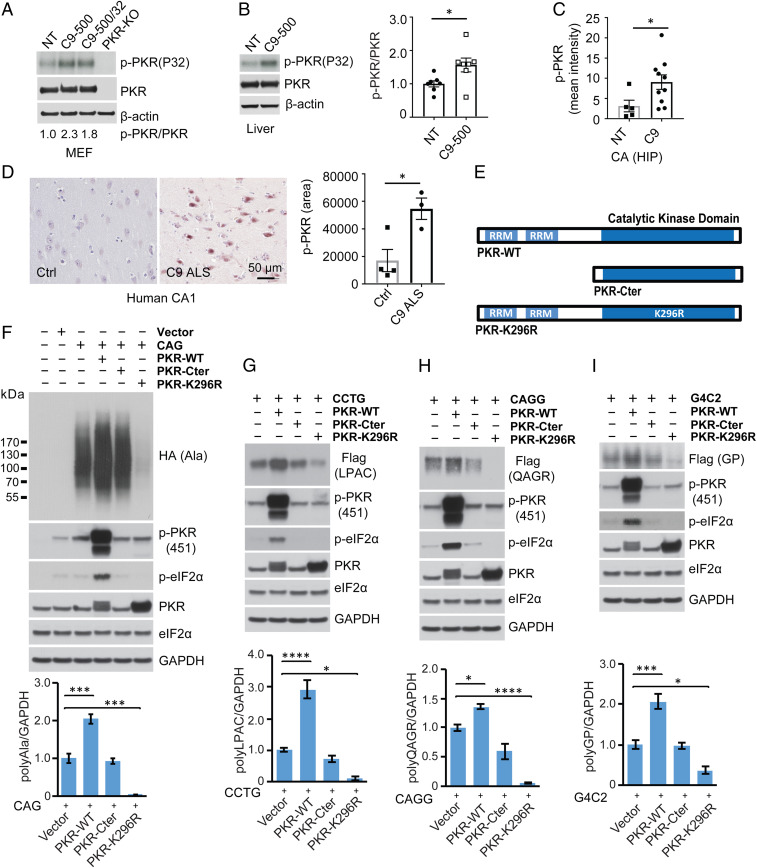
Fig. 1.
RAN translation of CAG, CCTG, CAGG, G4C2 expansions is regulated by PKR. (A and B) In vitro PKR kinase assay showing levels of p-PKR in MEFs (A) and liver (B) from C9 BAC and NT mice. (C) Quantification of p-PKR immunofluorescence staining in CA hippocampal region elevated in a cohort of phenotypic C9-500 vs. NT mice. (D) IHC showing α-p-PKR staining in CA1 of hippocampus is elevated in C9 human autopsy brains. (E) Schematic diagrams showing PKR-WT, PKR-C-terminal, and PKR-K296R constructs. (F–I) Immunoblots of HEK293T lysates after transfection with CAG (F), CCTG (G), CAGG (H), and G4C2 (I) expansion constructs with or without constructs expressing PKR-WT, PKR-CT, or PKR-K296R. Protein blots were probed for tagged RAN proteins, p-PKR, p-eIF2α, and total PKR. PKR overexpression increases phospho-PKR (p-PKR) and phospho-eIF2α (p-eIF2α), whereas PKR-Cter has no effect and PKR-K296R decreases steady-state levels of p-PKR and p-eIF2α. RAN protein quantification is shown in the lower graphs. Statistical analyses were performed using the t test (B–D) or one-way ANOVA with Dunnett analyses for multiple comparisons (F–I); *P < 0.05, ***P < 0.001, ****P < 0.0001; n ≥ 3 per group. Bars show mean ± SEM.
Next, we tested the effects of PKR overexpression and inhibition on RAN protein accumulation across several types of hairpin forming repeat expansions (19, 20). Repeat expansion plasmids were cotransfected with constructs expressing either full-length PKR (PKR-WT), an inactive PKR lacking the RNA binding domain (PKR-Cter), or a dominant negative form of the enzyme (PKR-K296R) that contains a catalytically inactive mutation in the ATP binding site (Fig. 1E). For CAG repeat-expressing cells, PKR-WT overexpression leads to increased p-PKR, p-eIF2α, and RAN polyAla protein levels expressed from the GCA reading frame (Fig. 1F). As expected, expression of the inactive form of the enzyme (PKR-Cter) had no effect on p-PKR, p-eIF2α, and RAN polyAla protein levels. In contrast, expression of the dominant-negative PKR-K296R mutation, harboring an inactive kinase domain, dramatically decreased polyAla RAN protein levels (Fig. 1F). Cells expressing CCUG, CAGG, and G4C2 repeats produce polyLeu-Pro-Ala-Cys (polyLPAC), polyGln-Ala-Gly-Arg (polyQAGR), and polyGly-Pro (polyGP) RAN proteins, respectively. Consistent with the polyAla results, LPAC, QAGR, and GP RAN protein levels increased with PKR-WT overexpression and decreased with PKR-K296R overexpression (Fig. 1 G–I and SI Appendix, Fig. S5A). No decreases in RNA levels were detected between treatment groups that could explain changes in RAN protein levels in PKR-WT–treated or PKR-K296R–treated cells (SI Appendix, Fig. S5 B–F). In contrast to the reductions seen in the polyAla reading frame, polyglutamine proteins expressed from CAG expansion constructs with or without an AUG-initiation codon were not similarly affected by PKR (SI Appendix, Fig. S6 A–C).
In summary, these data show that several types of hairpin-forming microsatellite expansion mutations activate the PKR pathway in vitro. Consistent with the in vitro data, p-PKR levels are increased in human and mouse tissues containing C9orf72 repeat expansions. Additionally, overexpression of PKR increases RAN protein expression and PKR inhibition dramatically decreases the levels of multiple types of RAN proteins in transfected cells.
RAN Translation Regulated by PKR Phosphorylation.
To directly test the role of PKR in RAN translation, we generated HEK293T PKR knockout (KO) cells using CRISPR/Cas9. Transfection experiments show that polyAla RAN protein levels are dramatically reduced in PKR-KO compared to control cells isolated after treatment with nontargeting gRNAs (Fig. 2A) and, as expected, PKR overexpression rescues polyAla RAN expression (SI Appendix, Fig. S7). Similar decreases in polyLPAC, polyQAGR, and polyGP RAN proteins were observed in the PKR-KO cells (Fig. 2 B–D).
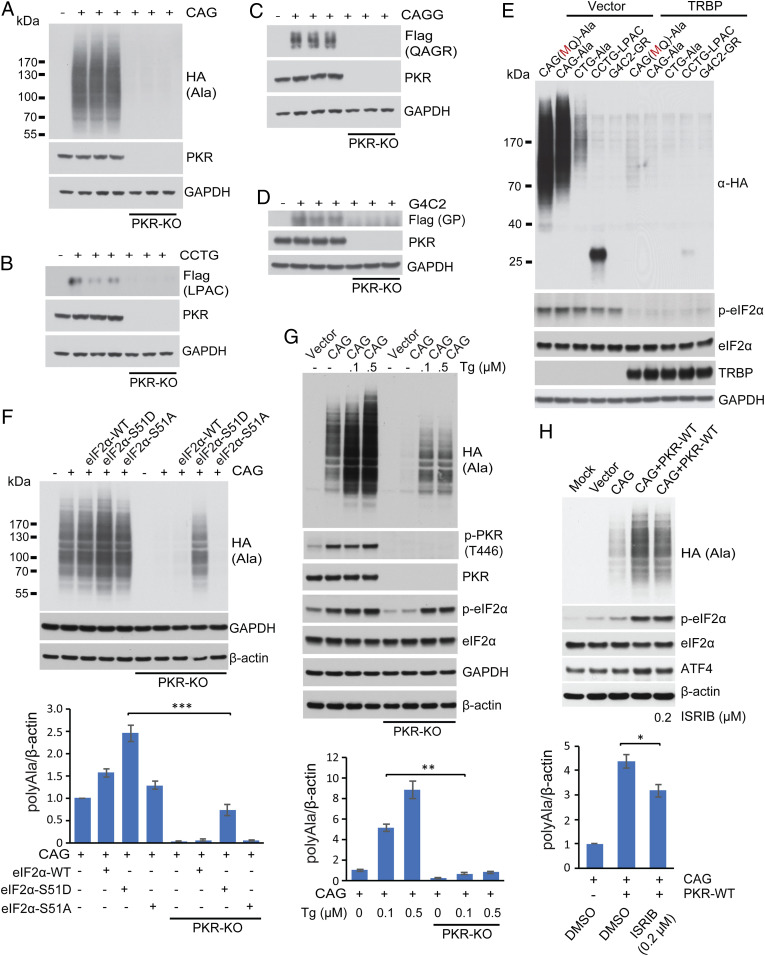
Fig. 2.
RAN protein levels regulated by PKR/eIF2α pathways. (A–D) Protein blots comparing polyAla (A), polyLPAC (B), polyQAGR (C), and polyGP (D) RAN protein levels in PKR-KO compared to control HEK293T cells. (E) Protein blots showing that overexpression of TRBP reduces levels of RAN polyAla expressed from CAG and CUG expansion RNAs and RAN polyLPAC expressed from CCUG repeats. Additionally, TRBP reduces levels of p-eIF2α. (F) Protein blots of HEK293T WT and PKR-KO cells after cotransfection with plasmids expressing a CAG expansion and eIF2α-WT, eIF2α-S51D, or eIF2α-S51A. Protein blots were probed with α-HA to detect polyAla RAN protein levels, which are quantitated in Lower. (G) Protein blots after thapsigargin treatment of WT and PKR-KO cells transfected with CAG repeat constructs with polyAla quantification. (H) Protein blots of ISRIB treatment of HEK293T cells cotransfected with CAG repeat and PKR-WT constructs with polyAla quantification. Statistical analyses were performed using the two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, n = 3 per group, Bars show mean ± SEM.
To further test the role of PKR in RAN translation, we inhibited PKR by overexpressing the PKR inhibitor TAR RNA binding protein (TRBP) (21, 22). Cotransfection experiments show TRBP overexpression decreases the levels of RAN polyAla expressed from CAG or CUG expansion RNAs and polyLPAC expressed from CCUG transcripts (Fig. 2E and SI Appendix, Fig. S8A) but not polyGln (SI Appendix, Fig. S8B). Consistent with a possible role of PKR in the integrated stress response pathway, PKR inhibition by TRBP decreases p-eIF2α (Fig. 2E).
Next, we performed a series of experiments to directly test if the p-eIF2α pathway increases RAN translation. We cotransfected HEK293T cells with CAG expansion constructs and plasmids expressing WT, the phosphomimetic eIF2α-S51D, or a nonphosphorylatable mutant eIF2α-S51A. These data show that the phoshomimetic eIF2α-S51D increased steady-state levels of polyAla RAN protein while the nonphosphorylatable eIF2α-S51A protein had no effect compared to WT eIF2α-expressing cells (Fig. 2F). Next, we examined whether p-eIF2α is sufficient to increase polyAla RAN protein levels in the absence of PKR. For this experiment, we cotransfected PKR-KO cells with CAG expansion plasmids and constructs expressing various forms of eIF2α (Fig. 2F). While these data show that expression of eIF2α-S51D is able to increase RAN polyAla levels in the absence of PKR, quantification shows that the levels of polyAla in WT cells were much higher than in PKR-KO cells (Fig. 2 F, Lower). Similarly, thapsigargin treatment, which causes eIF2α phosphorylation, via phosphorylation of PERK, only partially restores polyAla levels in PKR-KO cells (Fig. 2G). To further test the role of p-eIF2α in RAN translation, we inhibited p-eIF2α with the antagonist ISRIB in cells cotransfected with CAG repeat and PKR-WT constructs. Western blots show that polyAla levels were modestly reduced (∼25%) by ISRIB treatment in cells cotransfected with WT-PKR and a plasmid expressing CAG repeats (Fig. 2H).
Taken together, these results support a model in which chronic PKR activation by repeat expansion RNAs increase the levels of multiple types of RAN proteins. Additional experiments show that blocking PKR activation dramatically reduces RAN protein levels, whereas blocking the effects of p-eIF2α are less robust. These data suggest PKR affects RAN translation through both eIF2α phosphorylation-dependent and phosphorylation-independent pathways.
PKR-K296R Decreases RAN Proteins and Improves Behavior in C9orf72 ALS/FTD BAC Transgenic Mice.
To investigate whether PKR inhibition affects RAN protein levels in vivo, we performed rAAV-mediated delivery of dominant negative PKR-K296R or EGFP as a control via intracerebroventricular (ICV) injections at postnatal day 0 (P0) (Fig. 3A) C9orf72 BAC transgenic mice and nontransgenic (NT) littermate controls. Four groups of animals (EGFP/C9, EGFP/NT, PKR-K296R/C9, and PKR-K296R/NT) were aged to 3 mo, an age at which mice develop several early signs of disease. Southern blot analyses were performed as described in Liu et al. (15) to select groups of C9 (+) animals with comparable repeat lengths for further study (see SI Appendix, Fig. S9 for a typical example Southern blot). Immunohistochemistry (IHC) against the N-terminal V5 epitope tag on the PKR-K296R construct showed widespread transduction and expression of PKR-K296R throughout the brain (e.g., SI Appendix, Fig. S10A). As described (23), IF staining for EGFP after rAAV-EGFP P0 injections showed widespread transduction and EGFP expression in mouse brains at 3 mo of age (SI Appendix, Fig. S10B). Consistent with cell culture data, immunofluorescence staining using a p-eIF2α antibody shows C9 AAV EGFP-treated animals generally show more intense p-eIF2α staining compared to low levels seen in the AAV PKR K296R-treated mice (SI Appendix, Fig. S10 C and D). IHC using a monoclonal α-GA antibody (15) showed that C9 mice treated with PKR-K296R had an ∼50% decrease (P = 0.029) in polyGly-Ala (polyGA) aggregates in the aggregate-rich retrosplenial cortex compared to C9 EGFP controls (Fig. 3B). Because available antibodies worked better to detect soluble GP rather than aggregates in our mice, we used a meso scale discovery (MSD) immunoassay to measure soluble GP levels (24). Soluble GP levels were reduced by more than 60% (P = 4.06e-08) in brain lysates from C9 PKR-K296R–treated animals compared to C9 EGFP controls (Fig. 3C). C9 mice were previously shown to develop anxiety-like phenotypes by open-field analyses with reduced center time. Here, PKR-K296R–treated C9 animals showed improvement in open field testing with increased center time compared to C9 EGFP control animals (Fig. 3D). Additionally, DigiGait analyses, which quantitates specific features of gait dynamics and posture, showed that of the 15 parameters that differed between C9 EGFP-injected and NT EGFP-injected controls at 12 wk, nine parameters improved in C9 PKR-K296R–treated animals (Fig. 3E). Additionally, animals from the C9 PKR-K296R treatment and NT control groups showed similar performance by DigiGait with only 3 of 15 parameters significantly differing between them (Fig. 3F). Examples of two improved gait parameters in the C9 PKR-K296R treatment group are shown in Fig. 3 G and H.
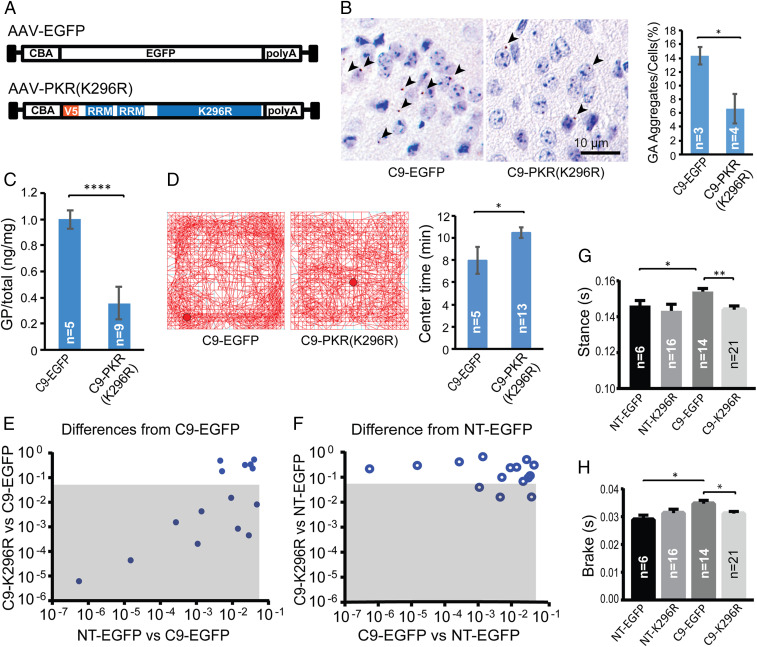
Fig. 3.
Inhibition of PKR decreases RAN proteins and improves behavior in C9-BAC mice. (A) Schematic diagrams of EGFP control and PKR-K296R AAV2/9 constructs used for ICV injections of C9-BAC and NT mice. (B) Representative IHC staining (brown color) of GA RAN protein aggregates in retrosplenial cortex from C9-EGFP and C9-PKR-K296R mice with quantification of GA RAN protein aggregates. Two-tailed t test; *P < 0.05, Bars show ± SEM. (C) MSD assay of brain lysates showing PKR-K296R–treated mice have lower levels of soluble GP RAN protein. Bars show ± SEM. (D) Open-field analyses of C9 mice treated with PKR-K296R or EGFP. (E and F) Comparisons of C9-relevant DigiGait parameters among AAV PKR-K296R and EGFP treatment cohorts at 3 mo of age. Gray boxes show parameters that significantly differ between PKR-K296R C9 and EGFP C9 cohorts (E) or PKR-K296R C9 and NT EGFP cohorts (F). (G and H) Example DigiGait data for Stance and Brake parameters. Statistical analyses were performed using two-tailed t tests (B–D), *P < 0.05, ****P < 0.0001 with corrections for multiple comparisons in G and H, *P < 0.025, **P < 0.005. Bars show ± SEM. All analyses were done in a blinded fashion.
In summary, C9 PKR-K296R–treated mice showed reduced levels of GA and GP RAN proteins and significant improvements in anxiety-like behavior and gait abnormalities compared to C9 EGFP-treated animals at 3 mo of age.
Metformin Decreases RAN Protein Levels and Mitigates Repeat Expansion-Induced PKR Activation.
Metformin is widely used for treating type 2 diabetes, and recent studies have shown it has beneficial effects in aging and neurodegenerative disorders (25, 26). In fragile X syndrome (FXS), metformin normalizes protein translation by inhibiting the MAPK/Erk pathway (27). Since metformin improved survival and behavioral phenotypes in Huntington disease mice (28, 29), a CAG microsatellite expansion disorder involving RAN protein accumulation (30), we tested the effects of metformin on RAN translation by expressing repeat expansion transcripts in cells treated with or without metformin. We found that metformin decreases polyAla, polyLPAC, and polyGP RAN protein levels (Fig. 4A), but not polyGln levels (SI Appendix, Fig. S11), in HEK293T cells expressing CAG, CCUG, or G4C2 expansion RNAs. Surprisingly, protein blots also show that metformin dramatically decreased PKR phosphorylation at the T446 and T451 sites, which are an indication of PKR activation (Fig. 4B). In contrast, only modest reductions in p-eIF2α levels were seen in metformin-treated cells (SI Appendix, Fig. S12). No changes in RNA levels were observed between metformin-treated and metformin-untreated cells (SI Appendix, Fig. S13). Metformin decreased p-PKR and p-eIF2α levels under the stress of DNA transfection (SI Appendix, Fig. S14A) (31, 32). Additionally p-PKR levels are reduced by metformin treatment in the presence of the small molecule PKR activator, 1H-benzimidazole-1-ethanol,2,3-dihydro-2-imino-α-(phenoxymethyl)-3-(phenylmethyl)-, monohydrochloride (BEPP) (SI Appendix, Fig. S14B) (33). Additionally, metformin and the related drugs phenformin and buformin show similar inhibition of G4C2 repeat expansion-induced p-PKR levels and RAN polyGP levels (SI Appendix, Fig. S15).
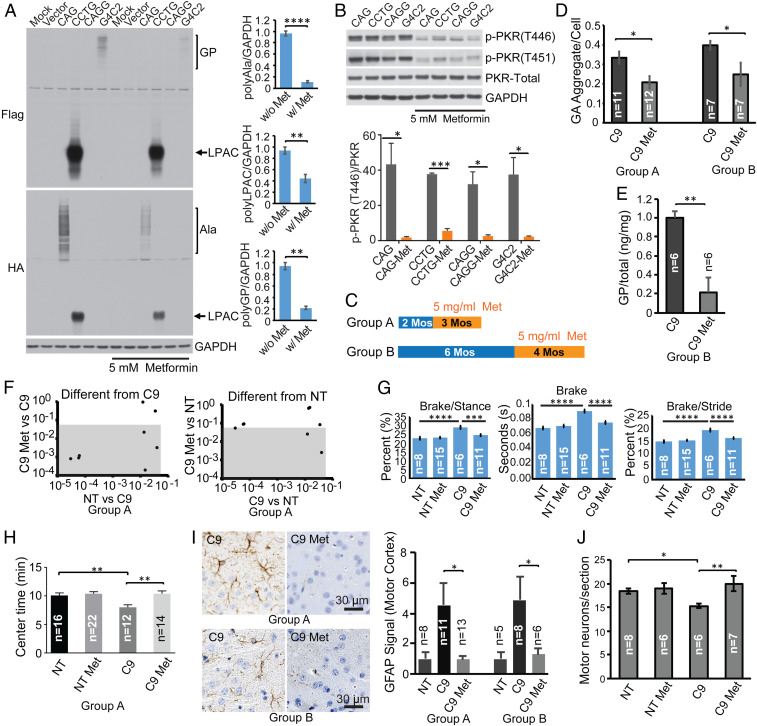
Fig. 4.
Metformin inhibits PKR, reduces RAN proteins, and ameliorates disease in C9-BAC mice. (A) Protein blots of HEK293T cells transiently transfected with CAG, CCTG, CAGG, and G4C2 expansion constructs (n = 3 per group) treated with or without 5 mM metformin. (B) Protein blots of total PKR, p-PKR (T446), and p-PKR (T451) from HEK293T cells transfected with various repeat expansion constructs with or without metformin treatment. (C) Animal study design with metformin treatment for 2–5 (group A) or 6–10 mos (group B). (D) Quantification of GA aggregates by IHC. (E) Soluble GP levels measured by MSD. (F) Comparisons of C9-relevant DigiGait parameters in cohorts with or without metformin treatment at 3 mo of age. Gray boxes show parameters that significantly differ between C9 cohorts with and without metformin (Left) or metformin C9 and NT control groups (Right). (G) Example data of three DigiGait parameters. (H) Open-field analyses. (I) GFAP staining in metformin-treated vs. untreated C9-BAC animals. GFAP levels were separately normalized to NT levels for group A and group B. (J) ChAT-positive motor neurons in the lumbar spinal cord (L3–L6) in C9-BAC and NT mice with or without metformin treatment from 2 to 10 mo. Statistical analyses were performed using two-tailed t test (A, F, and I), one-way ANOVA with Tukey analyses for multiple comparisons (G), and one-way ANOVA with Holm–Sidak analyses for multiple comparisons (H and J) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All analyses were done in a blinded fashion.
In summary, these data demonstrate that metformin reduces p-PKR and the levels of several types of RAN proteins in mammalian cells and identify metformin as a PKR inhibitor.
Metformin Ameliorates Neuropathological and Behavioral Phenotypes in the C9-BAC Mouse Model.
Next, we investigated whether metformin can decrease RAN protein levels in vivo and reverse behavioral phenotypes in C9-BAC mice. C9 and NT mice were treated with or without metformin. A dose of 5 mg/mL in the drinking water (Fig. 4C) was used in this study, which has previously been shown to result in plasma levels (∼10 µM) comparable to conventional human doses of 20 mg·kg−1·d−1 used in diabetic patients (34–36). In group A animals, 3 mo of treatment began at 2 mo of age, before the onset of overt behavioral or pathological phenotypes. In group B, a smaller cohort of animals were treated for 4 mo beginning at 6 mo, an age at which behavioral phenotypes are evident (15).
Molecular characterization showed that C9 metformin-treated animals had a 38% (group A, P = 0.01) and 37% (group B, P = 0.047) reduction in the number of GA aggregates in the retrosplenial cortex compared to C9 controls (Fig. 4D). Similarly, metformin-treated C9 animals had an ∼80% (P = 0.0036) decrease in levels of soluble GP compared to C9 controls in (group B) (Fig. 4E). In contrast, metformin did not change C9orf72 mRNA levels (SI Appendix, Fig. S16). Because GR aggregates are easier to detect in older animals, we extended metformin treatment in a cohort of animals beginning at 2 mo out to 18 mo of age. IHC in these animals showed a similar trend toward decreased GR aggregates with metformin treatment (SI Appendix, Fig. S17). Consistent with the cell culture work, metformin treatment decreased p-PKR levels in C9 BAC mice (SI Appendix, Fig. S18). p-eIF2α levels showed a trend toward reduction that did not reach statistical significance (SI Appendix, Fig. S19).
DigiGait analyses of group A mice at 5 mo identified eight parameters that differed between untreated C9 and NT cohorts. In C9 metformin-treated mice, six of these parameters improved compared to the C9 water treatment group (Fig. 4F) including brake, brake/stance, and brake/stride (Fig. 4G). Similarly, group A metformin-treated C9 mice showed increased center time by open field testing compared to untreated C9 mice (Fig. 4H). These data demonstrate that this anxiety-like behavior is improved by metformin treatment.
IHC staining for glial fibrillary acidic protein (GFAP), a marker of neuroinflammation previously reported to be up-regulated in the motor cortex of our C9-BAC mice (15, 24), was reduced by 79% (group A) and 74% (group B) in metformin-treated C9 animals compared to untreated C9 mice, and staining in C9-treated animals was comparable to NT animals (Fig. 4I and SI Appendix, Fig. S20).
Because motor neuron loss is a consistent feature of disease in older C9-BAC mice, and ALS patients, we treated an additional cohort of mice with metformin from 2 to 10 mo of age. Metformin treatment of C9 animals prevented motor neuron degeneration of the lumbar spinal cord that was observed in untreated C9 animals (P = 0.0087) (Fig. 4J and SI Appendix, Fig. S21).
Taken together, these data demonstrate that metformin reduces p-PKR and RAN protein levels in vivo, and metformin treatment improves behavior and decreases neuroinflammation and motor neuron loss in C9-BAC transgenic mice.
Discussion
We identify PKR as a key regulator of a pathway that controls the expression and accumulation of several types of RAN proteins and demonstrate that genetic or pharmacological inhibition of PKR reduces RAN protein levels. Additionally, we report that the FDA-approved drug metformin inhibits PKR phosphorylation, reduces RAN protein levels in vitro and in vivo, and mitigates disease in C9orf72 ALS/FTD BAC transgenic mice. These data identify PKR as a druggable target for RAN protein diseases and metformin as a promising FDA-approved drug that can be immediately tested in patients with C9orf72 ALS/FTD.
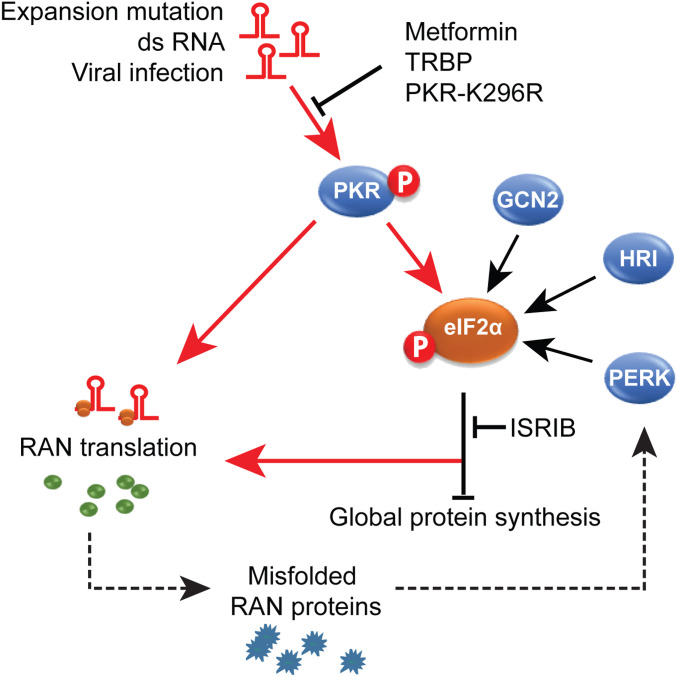
There are four kinases that phosphorylate eIF2α, which, in turn, leads to a general decrease of protein synthesis. Intense efforts have focused on targeting each of the kinases for various diseases including cancer, neurodegenerative diseases, and aging (37). Here, we demonstrate that the expression of four different hairpin-forming microsatellite expansion RNAs (19, 20) activate PKR and that PKR activation, leads to increased levels of a number of RAN proteins. Additionally, we show levels of p-PKR levels are elevated in C9orf72 human brains and C9-BAC transgenic mice. These data suggest a model in which repeat expansion RNAs lead to chronic activation of the PKR pathway, a condition that results in increased levels of p-eIF2α and the up-regulation of RAN translation (Fig. 5). Blocking the PKR pathway with PKR-K296R, metformin, or TRBP reduces RAN protein levels to a greater extent than blocking p-eIF2α with ISRIB. Additionally, stimulating the p-eIF2α pathway using eIF2α-S51D or thapsigargin in PKR-KO cells only partially rescues RAN protein expression. Taken together, these data suggest PKR regulates RAN protein levels through both p-eIF2α–dependent and p-eIF2α–independent pathways. The accumulation of misfolded or aggregated RAN proteins may, in turn, activate the PERK pathway and exacerbate disease by leading to further eIF2α phosphorylation and a cycle of increased RAN protein production (Fig. 5).
Fig. 5.
PKR activation model of RAN translation. Schematic diagram depicting chronic activation of PKR by repeat expansion RNAs increases RAN translation. Blocking the PKR pathway with PKR-K296R, metformin, or TRBP reduces RAN protein levels to a greater extent than blocking p-eIF2α with ISRIB. These results suggest PKR regulates RAN protein levels through both p-eIF2α–dependent and p-eIF2α–independent pathways. The accumulation of misfolded or aggregated RAN proteins may, in turn, activate the PERK pathway and exacerbate disease by leading to further eIF2α phosphorylation and a cycle of increased RAN protein production.
Our data show that several PKR inhibition strategies reduce RAN protein accumulation in mammalian cells: expression of dominant negative PKR-K296R, use of the PKR inhibitor TRBP, and knocking out PKR (Fig. 5). These data show that PKR activation is a major driver of RAN translation and identify the PKR pathway as a druggable target to reduce RAN protein expression. Furthermore, we show that blocking the PKR pathway in vivo decreases polyGP and polyGA RAN protein levels and improves behavioral and pathological phenotypes in C9-BAC mice. These data, combined with the lack of overt phenotypes and increased memory in PKR−/− mice (38) and therapeutic effects of PKR inhibition in Down syndrome (39), make PKR an attractive therapeutic target for C9orf72 ALS/FTD and other neurologic RAN protein diseases.
While PKR inhibition decreases the accumulation of several types of RAN proteins, (polyAla, polyLPAC, polyQAGR, and polyGP) in cell culture models, similar effects were not seen for polyGln expressed with or without an AUG-initiation codon. The fact that polyAla and polyGln are both translated from different reading frames of the same population of CAG expansion RNAs suggests that the effects of PKR on RAN protein accumulation can be frame specific. Additional studies that look at the synthesis and turnover of the polyAla versus polyGln proteins will be useful for understanding how PKR inhibition impacts the levels of RAN protein production in the polyAla and polyGln reading frames. In contrast, no similar frame specific effects were seen for polyLPAC accumulation, which was reduced in all three reading frames by PKR inhibition.
Metformin has been widely used as a safe, first-line defense against type 2 diabetes for decades. Although previous studies have shown metformin did not improve ALS in SOD1(G93A) overexpression mice (40), these mice do not have a repeat expansion mutation that produces RAN proteins and, therefore, these mice would not be expected to show improvements with metformin. In contrast, our data show metformin reduces RAN protein levels and improves ALS/FTD phenotypes in C9orf72 BAC transgenic mice containing a repeat expansion. When viewed together with our study, the previous observation that the incidence of ALS in type 2 diabetes patients is lower (41, 42) than in the general population suggests that metformin may protect a subset of ALS patients with the relatively common C9orf72 mutation, although further study is needed. Interestingly, metformin has shown beneficial effects in two other repeat expansion diseases. Huntington disease mice improve with metformin (28, 29), and Huntington disease patients that take metformin have been shown to score better on cognitive tests (43). Additionally, metformin improved mobility in patients with DM1 repeat expansions (44). Here, we describe a function of metformin as a PKR inhibitor. These data, combined with our C9-BAC mouse efficacy data, strongly support the notion of PKR inhibition and metformin as promising therapeutic strategies for C9orf72 ALS/FTD and possibly other RAN protein diseases.
Materials and Methods
Animal studies were approved by the Institutional Animal Care and Use Committee at University of Florida. Details of cell culture experiments, mouse experiments, metformin administration, AAV preparation, behavior, and molecular techniques are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Katsuya Nakamura for technical assistance and National Institutes of Health Grants RO1 NS098819, R37NS040389 and PO1-NS058901; Target ALS; ALS Association; Packard Center; Myotonic Dystrophy Foundation; Department of Defense Grant W81XWH1910654; and Muscular Dystrophy Association for support.
Footnotes
Competing interest statement: T.Z., L.N., and L.P.W.R. are listed as inventors on patents filed by the University of Florida related to RAN translation.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005748117/-/DCSupplemental.
Data Availability.
All other relevant data are described in the SI Appendix.
References
- 1.Cho D. H. et al., Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell 20, 483–489 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Moseley M. L. et al., Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 38, 758–769 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Zu T. et al., Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 108, 260–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ash P. E. et al., Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori K. et al., The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Zu T. et al., RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 110, E4968–E4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary J. D., Ranum L. P., New developments in RAN translation: insights from multiple diseases. Curr. Opin. Genet. Dev. 44, 125–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green K. M. et al., RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun. 8, 2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng W. et al., C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun. 9, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd T. W., Petrucelli L., Insights into the pathogenic mechanisms of Chromosome 9 open reading frame 72 (C9orf72) repeat expansions. J. Neurochem. 138 (suppl. 1), 145–162 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Taylor J. P., Brown R. H. Jr., Cleveland D. W., Decoding ALS: From genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edery I., Petryshyn R., Sonenberg N., Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: A novel translational control mechanism. Cell 56, 303–312 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Tian B. et al., Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 6, 79–87 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonenberg N., Hinnebusch A. G., Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y. et al., C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90, 521–534 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Nakamura T. et al., Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 140, 338–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bando Y. et al., Double-strand RNA dependent protein kinase (PKR) is involved in the extrastriatal degeneration in Parkinson’s disease and Huntington’s disease. Neurochem. Int. 46, 11–18 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Bañez-Coronel M. et al., A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 8, e1002481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dere R., Napierala M., Ranum L. P., Wells R. D., Hairpin structure-forming propensity of the (CCTG.CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J. Biol. Chem. 279, 41715–41726 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Ciesiolka A., Jazurek M., Drazkowska K., Krzyzosiak W. J., Structural characteristics of simple RNA repeats associated with disease and their deleterious protein interactions. Front. Cell. Neurosci. 11, 97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benkirane M. et al., Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 16, 611–624 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H. et al., TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc. Natl. Acad. Sci. U.S.A. 91, 4713–4717 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayers J. I. et al., Widespread and efficient transduction of spinal cord and brain following neonatal AAV injection and potential disease modifying effect in ALS mice. Mol. Ther. 23, 53–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen L. et al., Antibody therapy targeting RAN proteins rescues C9 ALS/FTD phenotypes in C9orf72 mouse model. Neuron 105, 645–662.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barzilai N., Crandall J. P., Kritchevsky S. B., Espeland M. A., Metformin as a tool to target aging. Cell Metab. 23, 1060–1065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gantois I., Popic J., Khoutorsky A., Sonenberg N., Metformin for treatment of fragile X syndrome and other neurological disorders. Annu Rev Med. 70, 167–181 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Gantois I. et al., Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nat. Med. 23, 674–677 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ma T. C. et al., Metformin therapy in a transgenic mouse model of Huntington’s disease. Neurosci. Lett. 411, 98–103 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Arnoux I. et al., Metformin reverses early cortical network dysfunction and behavior changes in Huntington’s disease. eLife 7, e38744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bañez-Coronel M. et al., RAN translation in Huntington disease. Neuron 88, 667–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kedersha N. L., Gupta M., Li W., Miller I., Anderson P., RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufman R. J., DNA transfection to study translational control in mammalian cells. Methods 11, 361–370 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Hu W. et al., Double-stranded RNA-dependent protein kinase-dependent apoptosis induction by a novel small compound. J. Pharmacol. Exp. Ther. 328, 866–872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y. et al., Antidiabetic drug metformin (GlucophageR) increases biogenesis of Alzheimer’s amyloid peptides via up-regulating BACE1 transcription. Proc. Natl. Acad. Sci. U.S.A. 106, 3907–3912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foretz M., Guigas B., Bertrand L., Pollak M., Viollet B., Metformin: From mechanisms of action to therapies. Cell Metab. 20, 953–966 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Memmott R. M. et al., Metformin prevents tobacco carcinogen–Induced lung tumorigenesis. Cancer Prev. Res. (Phila.) 3, 1066–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon S. L., Sonenberg N., Parker R., Neuronal regulation of eIF2α function in health and neurological disorders. Trends Mol. Med. 24, 575–589 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Zhu P. J. et al., Suppression of PKR promotes network excitability and enhanced cognition by interferon-γ-mediated disinhibition. Cell 147, 1384–1396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu P. J. et al., Activation of the ISR mediates the behavioral and neurophysiological abnormalities in Down syndrome. Science 366, 843–849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneb H. M., Sharp P. S., Rahmani-Kondori N., Wells D. J., Metformin treatment has no beneficial effect in a dose-response survival study in the SOD1(G93A) mouse model of ALS and is harmful in female mice. PLoS One 6, e24189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kioumourtzoglou M. A. et al., Diabetes mellitus, obesity, and diagnosis of amyotrophic lateral sclerosis: A population-based study. JAMA Neurol. 72, 905–911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jawaid A. et al., ALS disease onset may occur later in patients with pre-morbid diabetes mellitus. Eur. J. Neurol. 17, 733–739 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Hervás D. et al., Metformin intake associates with better cognitive function in patients with Huntington’s disease. PLoS One 12, e0179283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bassez G. et al., Improved mobility with metformin in patients with myotonic dystrophy type 1: A randomized controlled trial. Brain 141, 2855–2865 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other relevant data are described in the SI Appendix.