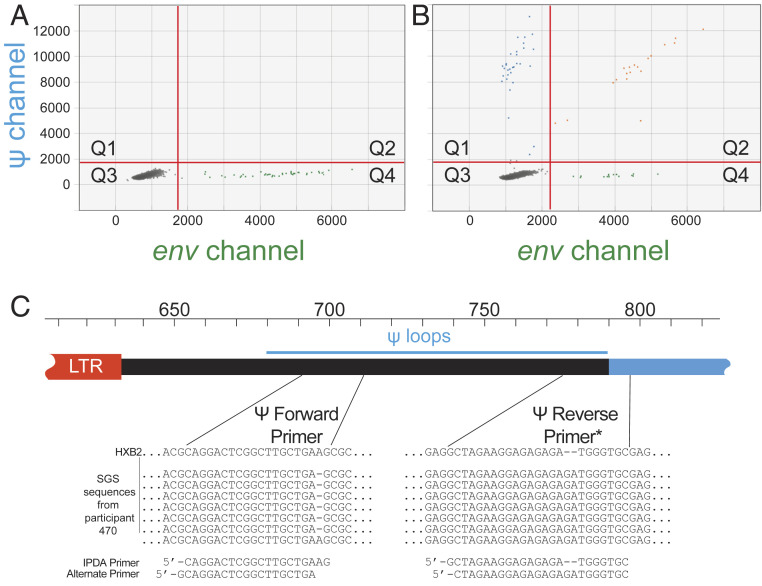
Fig. 4.
Custom IPDA oligos can resolve problematic mismatches, salvage amplification in the affected amplicon, and allow detection of intact proviruses. (A) IPDA dot plot showing ψ amplicon failure. (B) Restoration of the normal droplet pattern using custom primers based on single genome sequencing (SGS) of the ψ region. (C) SGS sequences of the ψ forward and reverse primer regions with custom primer design. A single nucleotide length polymorphism at the 3′ end of the ψ forward primer may have precluded amplification. There was also a two-nucleotide insertion in the ψ reverse primer. The use of alternate primers allowed successful ψ amplification shown in B. *Sequences are shown as sense strand nucleotides. The ψ reverse primers used in the IPDA are the reverse complements of the sequences shown.