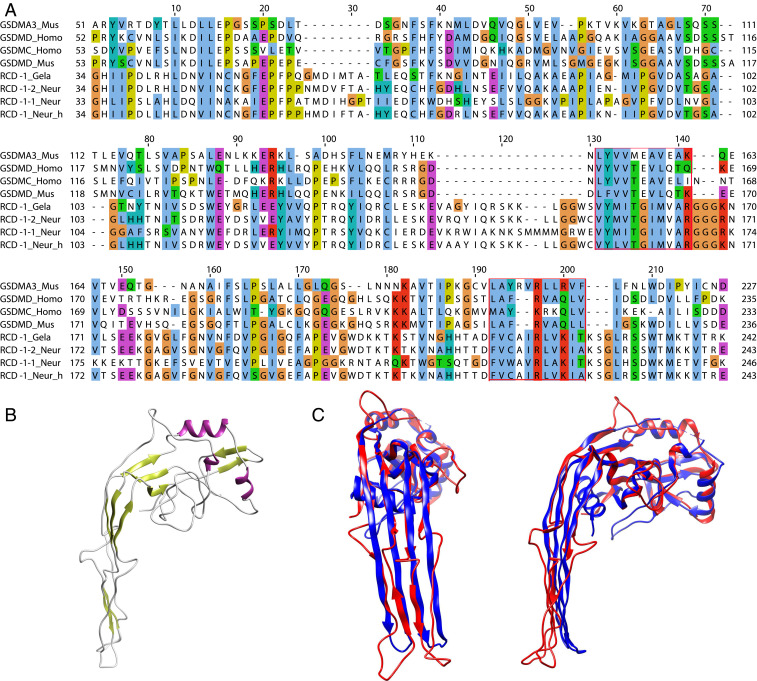
Fig. 1.
Protein alignment and molecular models of the pore-forming N-terminal domains of mammalian gasdermins and fungal RCD-1 proteins. (A) Alignment of protein sequences of murine (Mus) gasdermin-A3 (NP_001007462), gasdermin-D (NP_081236), human (Homo) gasdermin-D (NP_079012), gasdermin-C (NP_113603) with RCD-1–1 (XP_962898) and RCD-1–2 from N. crassa (Neur) and RCD-1 variants from N. hispaniola (Neur_h) and Gelasinospora tetrasperma. Conserved residues are shown in ClustalX colors, based on the nature of the amino acid residues: hydrophobic – blue, polar – green, negative charge – magenta, positive charge – red, aromatic – cyan, glycine – orange, proline – yellow, cysteine – pink, any/gap – white. Two highly conserved blocks between all sequences are boxed in red. (B) Template-based model of the RCD-1–2 allelic variant from Neurospora crassa. Murine GSDMA3 (6CB8) in the active pore-forming state was determined (P value = 7.22e-05) as the best template based on the sequence input (RCD-1–2). (C) Structural alignment of GSDMA3 (6CB8A) (blue) and the generated model of RCD-1–2 (red). Shown are a frontal view (Left) and side view (Right) of the two overlaid structures.