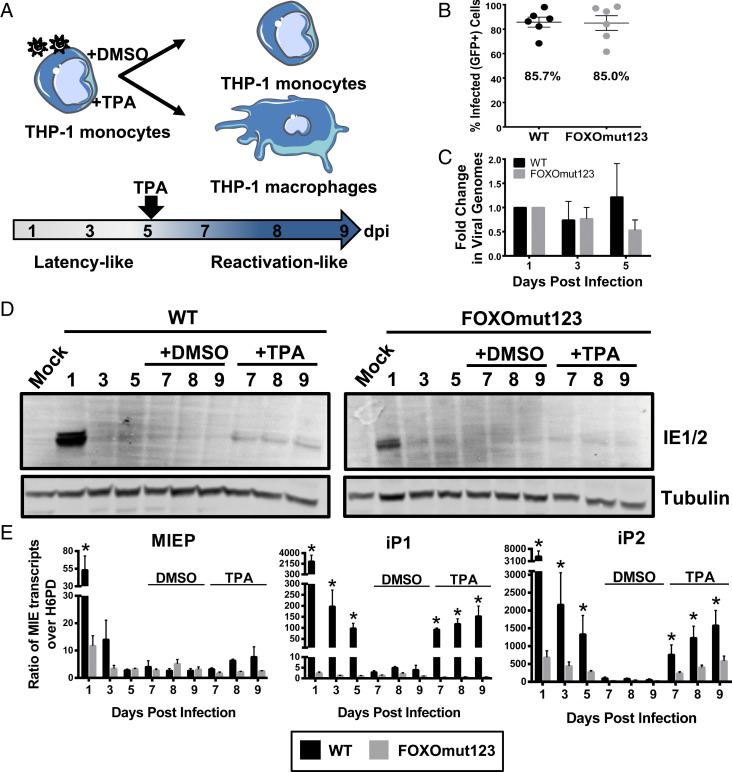
Fig. 5.
FOXO consensus sites are required for reexpression of MIE genes in THP-1 cells. (A) Schematic of THP-1 model for reexpression of MIE genes after stimulation with TPA. THP-1 cells were infected with TB40/E WT or FOXOmut123. At 5 dpi, cells were treated with TPA to stimulate macrophage differentiation and reexpression of viral genes, or with a DMSO vehicle control. (B) Equal infection (GFP+) of THP-1 cells with each virus was determined by flow cytometric analysis at 24 h postinfection (hpi). Biological replicates are represented as single points around the mean. SE is shown. (C) Total DNA was isolated and viral genomes were quantified using a primer to the β2.7 region of the HCMV genome relative to a cellular gene, RNase-P. Ratio of viral-to-cellular DNA at 3 and 5 dpi are normalized to their respective 1 dpi. Data from five independent biological replicates are shown; SEM is depicted. Two-way analysis of variance (ANOVA) showed no statistically significant change in genome copy for either infection over the time course. (D) Protein lysates were harvested at the indicated time points and immunoblotted to detect viral IE1 and IE2 with tubulin as a loading control. (E) MIE transcripts derived from the MIEP, iP1, or iP2 were quantified using reverse transcriptase quantitative PCR relative to the low-abundance housekeeping gene H6PD. Error bars represent the average of three independent experiments amplified in triplicate and SEM is shown. Statistical significance (*P ≤ 0.05) was determined by multiple paired t tests comparing accumulation of each transcript in WT versus mutant virus infection at each time point.