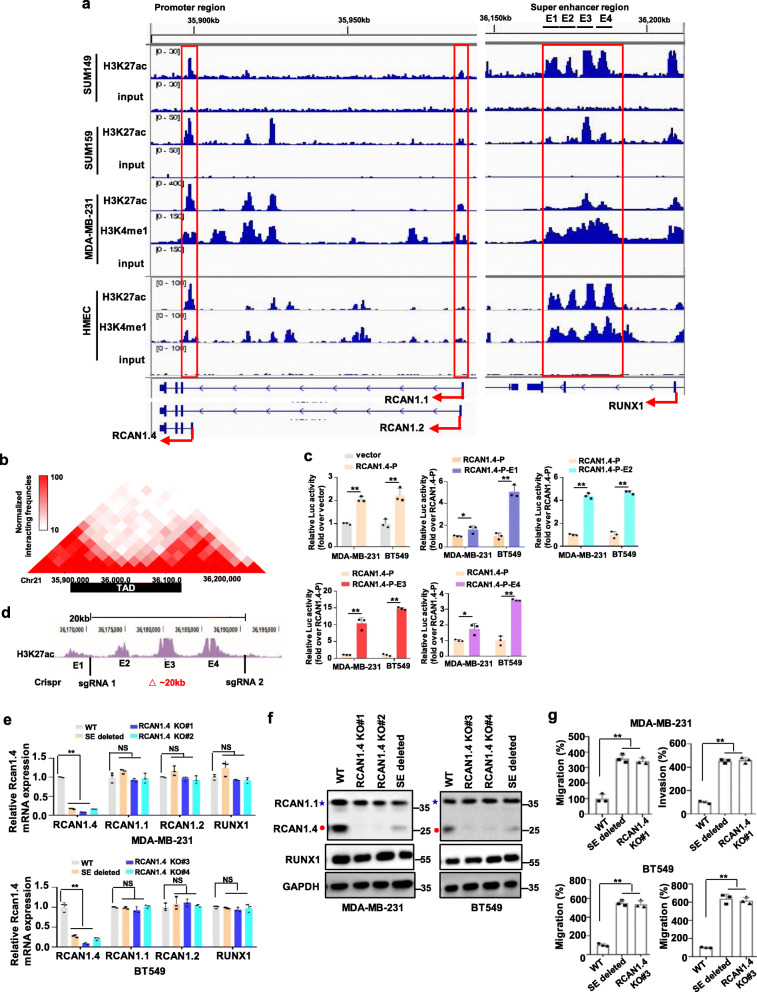
Fig. 3.
RCNA1.4-SE distal is responsible for the expression and function of RCNA1.4. a Gene tracks depicting the promoter/super-enhancer region of RCAN1.4 and the promoter region of RCAN1.1/RCAN1.2 in mammary epithelial cell HMEC and multiple breast cancer cells with measured H3K27ac, or H3K4me1 marks. The data were retrieved from Encode project (HMEC), GSE63581(SUM149 and SUM159) and GSE72141(MDA-MB-231). b RCAN1.4 in HMEC cell line topologically associated domain (TAD) region was predicted on the basis of the Hi-C data (http://promoter.bx. psu. Edu/ hi-c/view.php). c The luciferase activities of four enhancer elements were measured through Dual-Luciferase Reporter Assay in MDA-MB-231 and BT549 cells. d Schematic representation of deleting distal super-enhancer region of RCAN1.4 strategy. The location of two small guide RNA flanking the 20 kb RCAN1.4-SEdistal was shown. e-f Deletion of the human composite RCAN1.4 super-enhancer region in MDA-MB-231 and BT549 cells regulated the expression of the indicated genes. The mRNA levels were quantified using qRT–PCR(e). The cell lysates were prepared for immunoblots (f). Blue star, RCAN1.1; Red closed circle, RCAN1.4. g Deletion of the human composite RCAN1.4 super-enhancer region affected the migration and invasion ability of MDA-MB-231 and BT549 cells. Quantification of migratory and invasive cells of images per group was shown. Error bars represent mean ± SD, n = 3 biological independent samples. NS, not significance, * P < 0.05, **P < 0.01. The P value in e, g was determined by one-way analysis ANOVA with Dunnett’s multiple comparisons test, no adjustments were made for multiple comparisons. The P value in c was determined by a two-tailed unpaired Student’s t test. Data are representative of three independent experiments