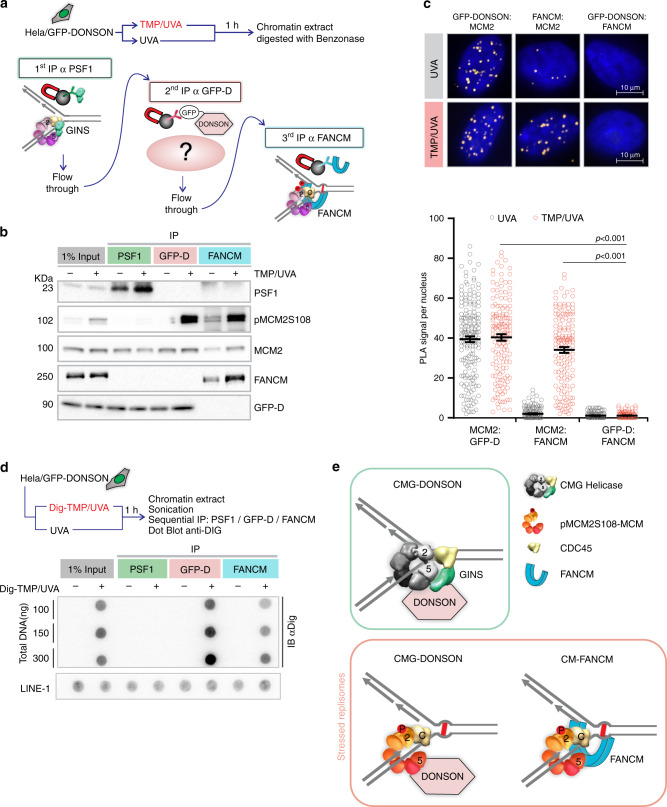
Fig. 2. DONSON and FANCM are on different replisomes.
a Scheme of sequential IP against DONSON and FANCM-associated replisomes. HeLa cells expressing GFP-DONSON were exposed to UVA only or TMP/UVA. Chromatin was prepared and digested with benzonase. This was followed by IP against PSF1 (to remove unstressed replisomes), then IP of the supernatant against GFP (to remove remaining DONSON-associated proteins), and finally IP of the residual supernatant to capture FANCM-bound proteins. b Western blot analysis of sequential IP. Representative blot (n = 3). c PLA in cells exposed to UVA only or TMP/UVA shows interactions between GFP-DONSON and MCM2; and FANCM and MCM2; but not between GFP-DONSON and FANCM. Scored nuclei: PLA between GFP-D: MCM2, UVA treatment = 174; TMP/UVA = 148; PLA between FANCM: MCM2, UVA = 142; TMP/UVA = 145; PLA between GFP-D: FANCM, UVA = 135; TMP/UVA = 133 from three biological replicates. Data are mean ± s.e.m. A two-sided Mann–Whitney rank-sum test was used to determine if differences were significant. NS: not significant: P > 0.05. d Association of replisomes with Dig-tagged ICLs. Chromatin was prepared from cells exposed to UVA or Dig-TMP/UVA, and the DNA reduced to fragments of <500 bp by sonication. Sequential IP was performed, and the DNA isolated from each fraction, dotted onto the nitrocellulose, and probed with an antibody to the Dig tag. LINE-1 repeat element served as a loading control. Representative blot (n = 2). e Model summarizing the results of the sequential IP experiment. Source data are provided as a Source Data file.