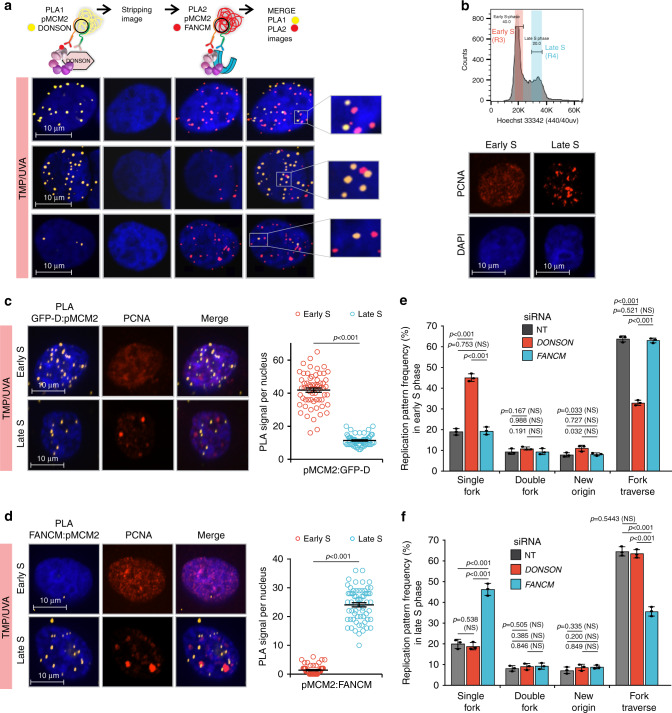
Fig. 3. DONSON and FANCM replisomes are active in different stages of the S phase.
a Analysis by sequential PLA of GFP-DONSON: pMCM2S108 complexes and then FANCM: pMCM2S108, in GFP-DONSON expressing cells exposed to TMP/UVA. After the first PLA, the cells were photographed (first column of images) and the antibodies and PLA product stripped (second column). The second PLA was performed, and the cells re-imaged (third column). The fourth column shows a merge of both images after image registration in the xyz planes using the DAPI signal. Examples are shown of cells with strong signals from both first and second PLA, or strong signals from the first but infrequent from the second, or weak from the first and strong from the second. The signals from the two PLA do not colocalize. b Early and late S phase fractions were isolated from sorted cells. The PCNA staining pattern from each fraction. c GFP-D: pMCM2S108 PLA in sorted early and late S phase cells. Scored nuclei: GFP-D: pMCM2S108 of early S phase = 62, late S phase = 60, from three biological replicates. Data are mean ± s.e.m. d FANCM: pMCM2S108 PLA in sorted early and late S phase cells. Scored nuclei: FANCM: pMCM2S108 of early S phase = 63, late S phase = 64, from three biological replicates. Data are mean ± s.e.m. e, f Influence of DONSON and FANCM on patterns of replication encounters with ICLs in early and late S phase cells. Cells were treated with siRNA against DONSON or FANCM, exposed to Dig-TMP/UVA and pulsed with nucleoside analogues as in Fig. 1a. Cells were sorted, and fiber patterns from early and late S phase analyzed. Data are mean ± s.d.. For PLA experiments (c, d), a two-sided Mann–Whitney rank-sum test was used, for replication pattern experiments (e, f) a two-sided unpaired t test was used, to determine if differences were statistically significant. NS: not significant: P > 0.05. Source data are provided as a Source Data file.