Fig. 3. Network of individuals sharing IBD segments over SOD1.
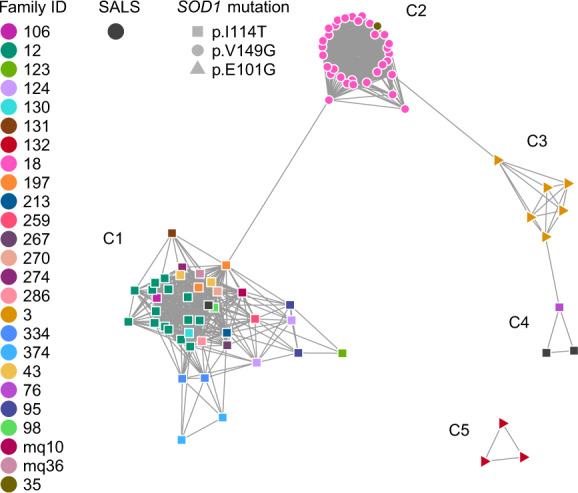
Each node is a sample and an edge was drawn between two samples if they were inferred IBD over SOD1. Nodes are positioned according to the Fruchterman–Reingold force-directed layout39, where there is no meaning behind the edge lengths. Nodes are coloured according to their unique family ID, in addition to the three sporadic ALS cases who have been assigned one colour. All samples had one of three SOD1 mutations, represented by unique node shapes in the network. There are five clusters in this network, denoted C1–C5, where all cases within each cluster had an identical SOD1 mutation. Cluster C2, where individuals carried SOD1 p.V149G (c.446T>G), connects family 18 and family 35, indicating they were in fact one family. Similarly, two clusters are present for individuals that carried SOD1 p.I114T (c.341T>C) (clusters C1 and C4), where these individuals were from different families, including three apparently sporadic ALS cases, indicating two disjoint extended families. Specifically, two sporadic cases were found to be related to each other and family 76 (cluster C4), whereas the third sporadic case was found to be related to the remaining 20 families with SOD1 p.I114T (cluster C1). In contrast, SOD1 p.E101G (c.302A>G) was unique to each family with this mutation (clusters C3 and C5), suggesting independent origins. The three pairs of individuals with discordant mutations who were inferred IBD over SOD1 did not share the disease-associated haplotypes and likely represent false-positive IBD calls.
