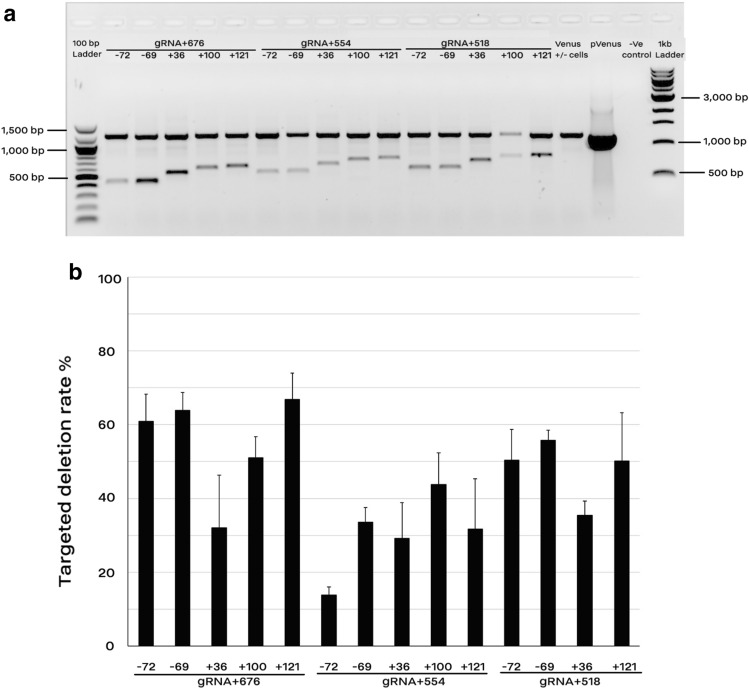
Figure 5.
Targeted deletions of Venus transgene by co-electroporation of two gRNA-encoding plasmids. (a) End-point PCR showed deletion of 15 different fragments from the original amplicon (1204 bp) using pairwise combination of gRNAs targeting the promoter/ 5′ region (− 72, − 69, + 36, + 100, and + 121) and the 3′ region (+ 518, + 554, and + 676) of Venus cDNA. The primer set (Venus-Forward1 and Venus-Reverse2) amplified both the shortened and original fragments. Genomic DNA from MEF cells carrying no-copy of Venus was considered as the negative controls (− Ve control). (b) Digital-PCR results for quantification of the rate of targeted deletion via a primer–probe assay. The primer–probe assay was designed to amplify only the wild type fragment. The deletion rate was calculated as the rate of copy number/µl in the co-electroporated groups divided by that of their control (non-treated) counterparts. Because of a poor DNA quality, the combined group of gRNA + 518 and + 100 was removed from the digital PCR analysis. Data are depicted as average ± standard error (n > 3).