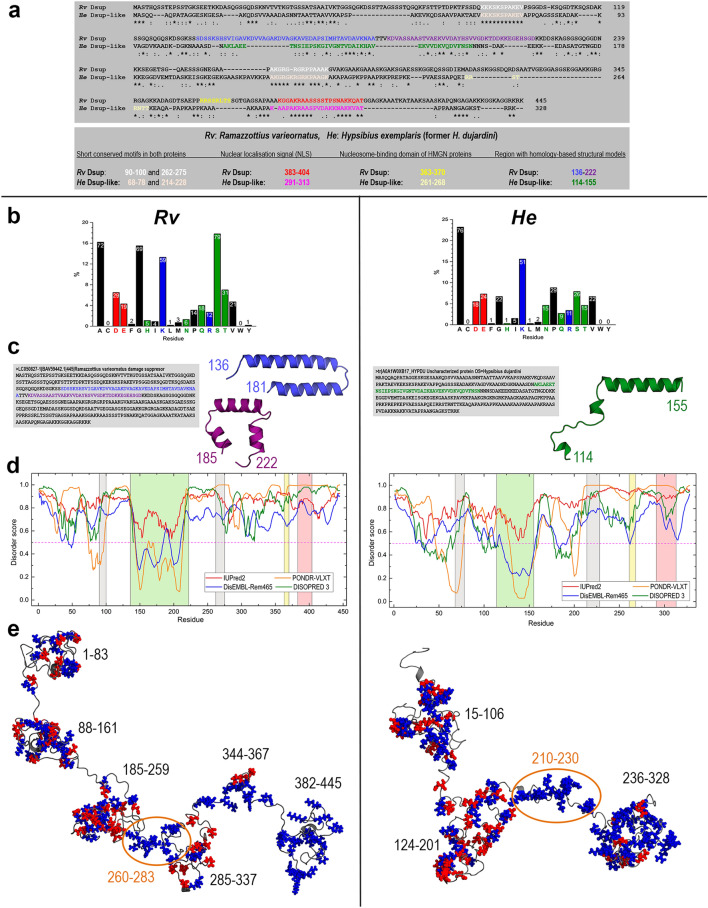
Figure 1.
Sequence and structural features of Dsup and Dsup-like proteins. (a) Sequence alignment obtained with Clustal-Omega 1.2.4 of the 445-residue Dsup protein from R. varieornatus (Rv) and the 328-residue Dsup-like protein from H. exemplaris (He) (“*” indicates perfect alignment whereas “:” and “.” indicate sites belonging to groups showing strong and weak similarity, respectively), highlighting the sequence motifs indicated in the box below. (b) Bar chart giving the % and number of each amino acid type according to the colour code: black = non-polar, green = polar, blue = positively charged, red = negatively charged. (c) Sequence segments with their corresponding structural models obtained by homology-modelling with SwissModel. (d) Structural disorder predicted from sequence with four predictors. The disorder score is defined in the range 0–1 with values > 0.5 meaning disorder (DISOPRED 3 gives no values with score < 0.5). Conserved motifs in both proteins, segments modelled by homology-modelling, HMGN-like and NLS sequences are the regions shaded light grey, light green, light yellow and light red, respectively. (e) Structural 3D models predicted from sequence by I-TASSER. Charged amino acids are shown as balls (positive in blue and negative in red) with labels indicating the sequence segments in which they accumulate. Regions marked with orange ovals enclose the second conserved motif 262–275 and 214–228 in Rv and He proteins, respectively.