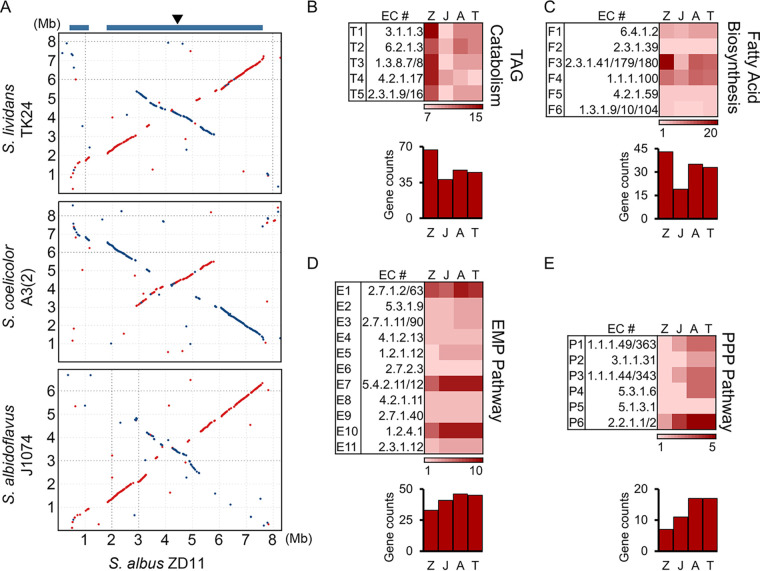
FIG 2.
Genomic landscape of S. albus ZD11. (A) Whole-genome alignments between S. albus ZD11 and S. albidoflavus J1074, S. coelicolor A3(2), and S. lividans TK24. Matches on the same strand are in red, and those on the opposite strand are in blue. The blue bars at the top represent the conserved core regions on the S. albus ZD11 chromosome. The arrowhead indicates the position of oriC. (B through E) Quantities of genes involved in the catabolic pathways for TAGs (B), the FA biosynthetic pathway (C), the EMP pathway (D), and the PPP (E). The quantity variance of genes corresponding to the KEGG pathway as determined for each biochemical reaction (k number) is color coded, as shown at the bottom of each pathway chart. Z, J, A, and T represent S. albus ZD11, S. albidoflavus J1074, S. coelicolor A3(2), and S. lividans TK24, respectively.