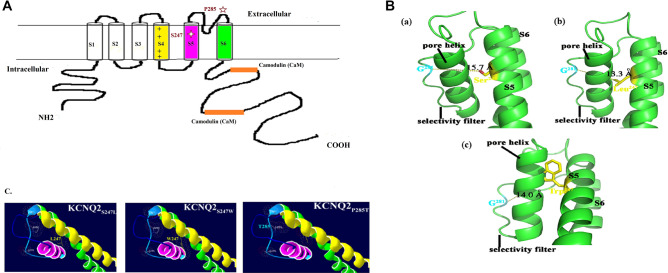
Figure 2.
(A) The schematic representation of KCNQ2 subunit with the position of the mutations of S247L, S247W and P285T. (B) Molecular modelling of KCNQ2 channel proteins (NM_004518) was generated by using the Phyre2 tool (Protein Homology/analogY Recognition Engine V 2.0) with the NP_004509.2 protein sequence. This tool is available on a website for protein modelling, prediction, and analysis based on the CryoEM structure of the Xenopus KCNQ1 channel54. The predicted 3D model of the KCNQ2 channel protein (c5vmsA_.1.pdb) was then used to analyse the structural difference between WT and mutant cells by using Swiss-PDBV (4.1.0 https://spdbv.vital-it.ch/) and PyMOL (https://www.pymol.org/), respectively. (a) WT S247, (b) S247L mutation, and (c) S247W mutation. The distance between residue 281 (G) on the selectivity filter and residue 247 on S5 of KCNQ2 was estimated as 15.7 Å for the WT (S247), 13.3 Å for the S247L mutant, and 14 Å for S247W mutant. Ion accessibility through the channel pore decreased for both mutants (S247L and S247W) at the S5 segment with bulkier side chain replacements compared with the WT (S247). (C) Mutation sites at the selectivity filter (P285T) might alter accessibility for potassium ions through the channels. Yellow color indicates S4; pink, S5; green, S6.