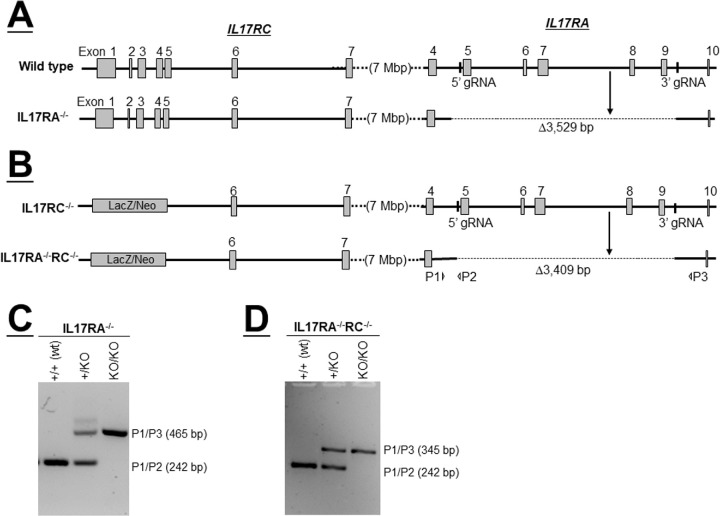
Figure 1.
Generation of IL17RA−/− and IL17RA−/−RC−/− mice. (A) Schematic diagram of IL17RA−/− mice. Top diagram depicts genomic loci of IL17RC and IL17RA in wt mice. Vertical bars show binding sites of guide RNAs (gRNAs) used to target Cas 9 to exons 5 and 9 of IL17RA using wild type mice (exons are shown as gray boxes). Deletions generated by CRISPR method are shown as dotted lines in the bottom diagram; (B) Schematic diagram of IL17RA−/−RC−/− mice. Top diagram shows genomic loci of IL17RC and IL17RA in IL17RC−/− mice. Similar to A above, gRNAs were used to delete exons 5 to 9 of IL17RA in IL17RC−/− mice. Deletions generated by CRISPR method are shown as dotted lines in the bottom diagram, and P1, P2, and P3 indicate binding sites of primers used to detect deletions by PCR; (C) PCR to detect deletion of IL17RA in wt mice. DNA from mice generated in A was used to confirm deletion of IL17RA. PCR primers P1 and P2 yield the PCR product from wild type allele, while PCR primers P1 and P3 yield PCR products from the knockout alleles; and (D) PCR to detect deletion of IL17RA in IL17RC−/− mice. DNA from mice generated in B was used to confirm deletion of IL17RA in IL17RC−/− mice. PCR primers P1 and P2 yield the PCR product from wild type allele, while PCR primers P1 and P3 yield PCR products from the knockout alleles.