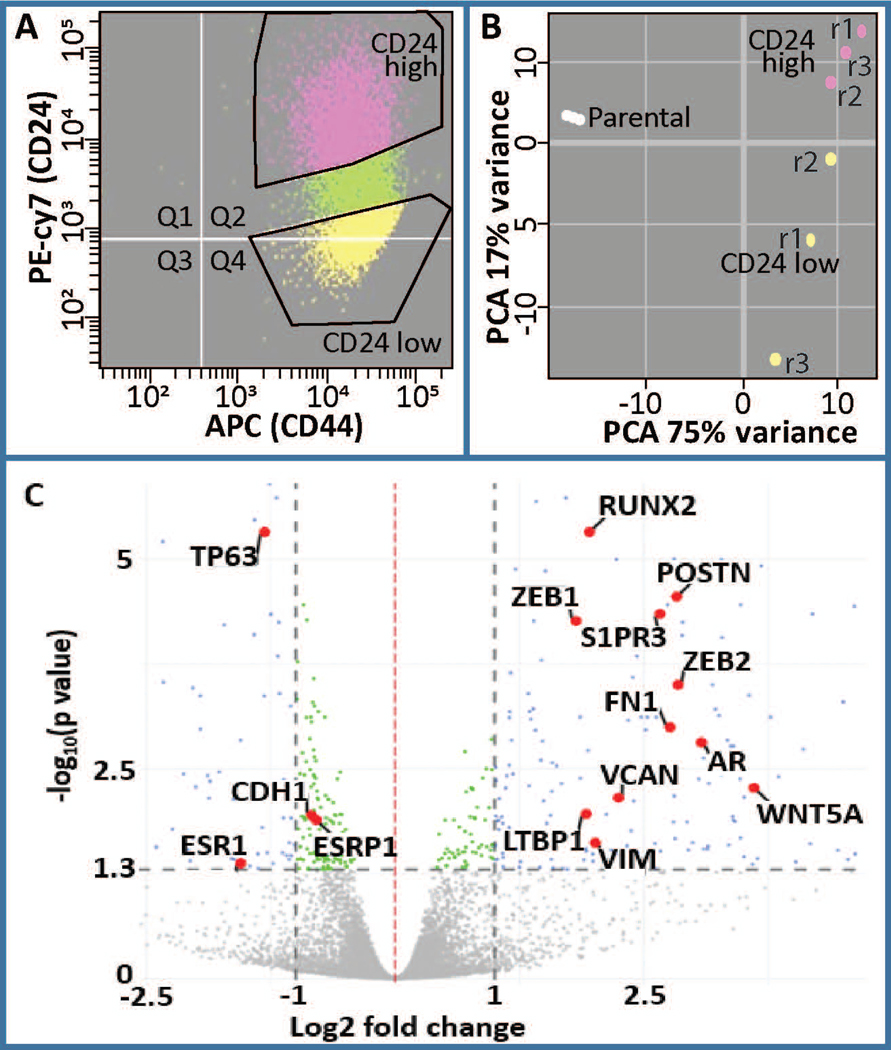
Figure 1.
Analysis of differentially expressed genes in CD24-/low/CD44+/high BCSCs versus CD24+/high/CD44high premalignant basal MCF10AT1 breast cancer cells. (a) FACS was performed on MCF10AT1 using CD24 (PE-cy7) and CD44 (APC) antibodies to isolate BCSCs (CD24−/low/CD44+/high, yellow) and bulk non-BCSC (CD24+/high/CD44+, pink). r1, r2, r3 refer to different replicates of FACS. (b) Principle component analysis demonstrated clustering of these two populations from each other. Parental cells were not subjected to FACS therefore suggesting an affect of FACS on gene expression. (c) A volcano plot demonstrating differentially expressed genes highlighting several genes that are well known to be altered in BCSCs including RUNX2, AR, ESR1, POSTN (Morra & Moch, 2011), CDH1, ESRP1, VIM, ZEB1, ZEB2, LTBP1, VCAN, WNT5A, and S1PR3 (Milara et al., 2012). BCSC, breast cancer stem cell; FACS, fluorescence-activated cell sorting; PCA, principal component analysis