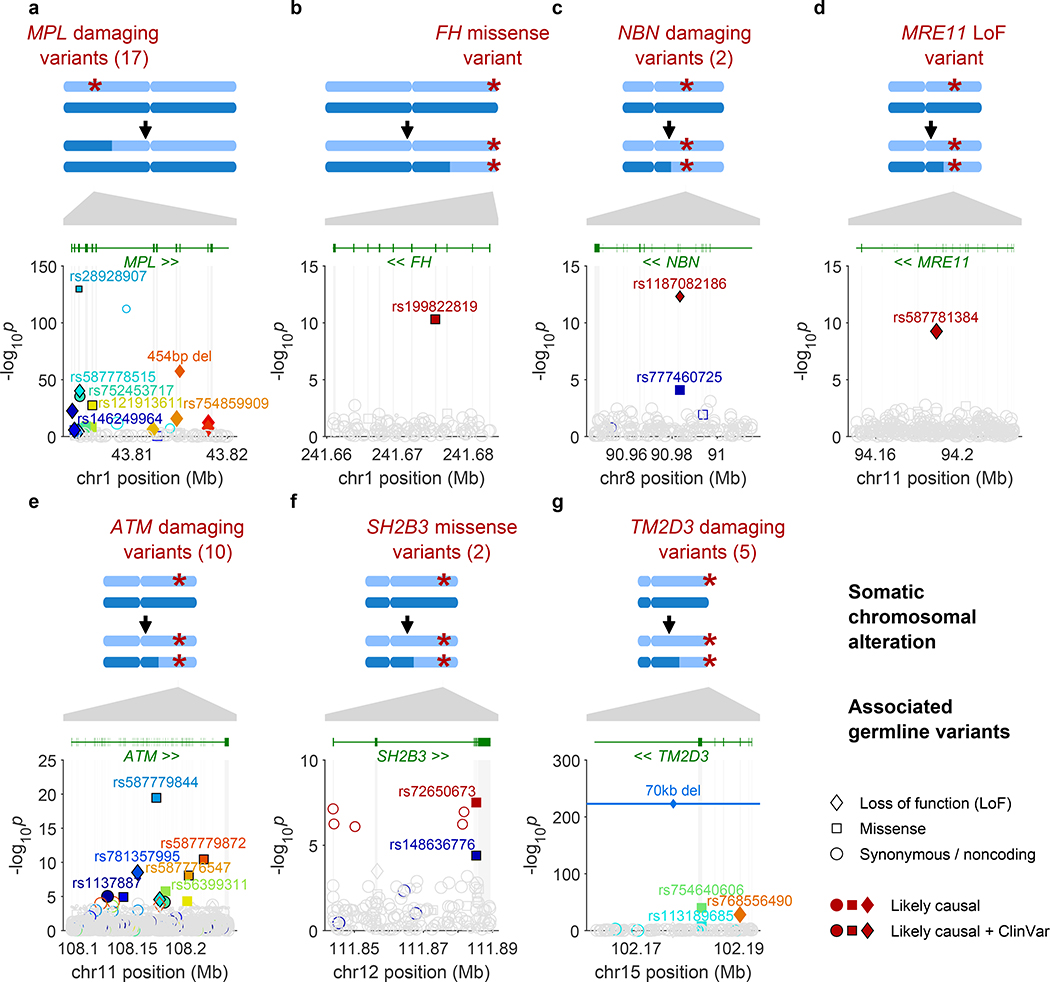
Figure 1: Fine-mapped inherited sequence alleles associated with the acquisition/selection of CN-LOH mutations in cis.
(a) MPL, (b) FH, (c) NBN, (d) MRE11, (e) ATM, (f) SH2B3, (g) TM2D3. At each locus, the CN-LOH mutations acquired by expanded clones tend to have deleted (a) or duplicated (b–g) the inherited alleles in a predictable manner as shown. Each panel is organized in the following way: top, genomic modifications observed in clones; bottom, association P-values (two-sided Fisher’s exact test on n≥378,307 individuals; Methods) vs. chromosomal position. All variants with filled symbols are likely causal coding or splice variants (Extended Data Table 1); black marker edges indicate evidence of pathogenicity in ClinVar21. Distinct colors are used to indicate the statistical independence of variants; any variants in linkage disequilibrium with likely causal variants (R2>0.2 in cases) are indicated with open symbols with a border color matching that of the likely causal variant. Symbol shapes indicate the effects of the indicated variant on encoded protein (LoF, missense, etc.); symbol sizes scale inversely with minor allele frequency.