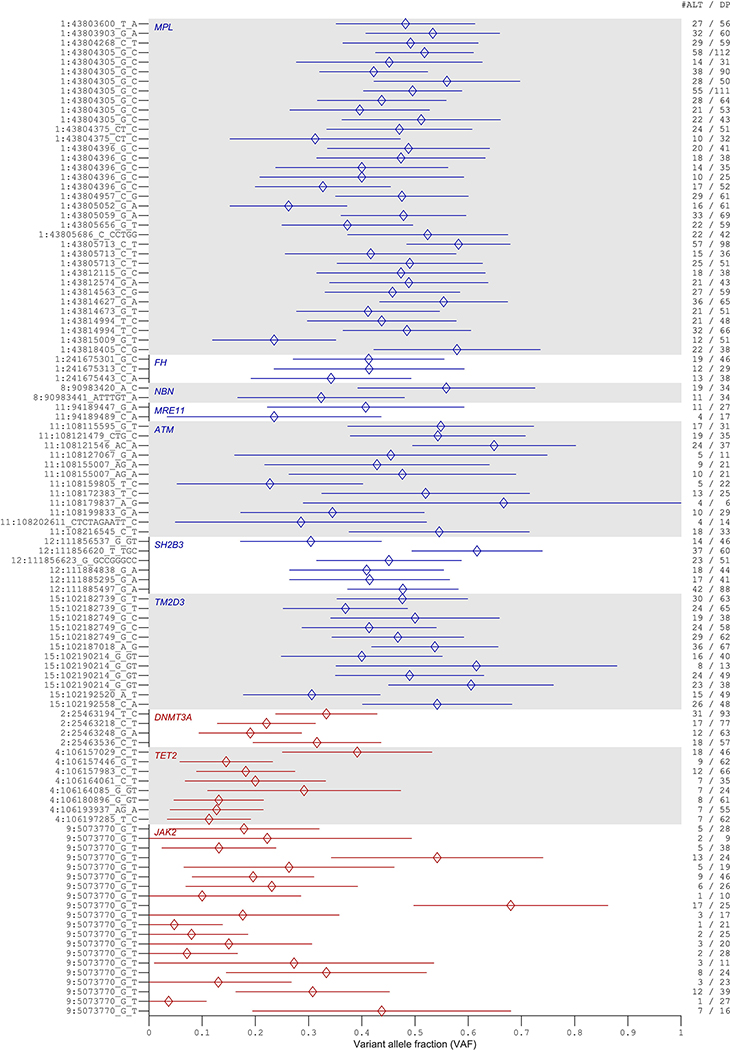
Extended Data Figure 8: Variant allele fractions of rare coding or splice variants likely to be targets of CN-LOH mutations in exome-sequenced individuals.
Variant allele fractions (VAF = number of reads matching the alternate allele divided by the total number of reads matching either the reference or the alternate allele) are plotted for each variant call identified as the potential target of a CN-LOH event (either from association analyses or burden analyses). Error bars, 95% CIs approximated using binomial standard errors multiplied by 1.96. Allelic read depths for variants identified at DNMT3A, TET2, and JAK2 are broadly indicative of somatic origin (VAF<0.5), while read depths for variants at the seven inherited risk loci are broadly consistent with inherited variation (VAF≈0.5). Read depths were generally insufficient to make a confident assessment of somatic vs. inherited origin on a per-variant level, as evidenced by wide VAF error bars; additionally, making this determination is further complicated by mapping bias toward the reference allele, which can produce VAF lower than 0.5 even for inherited variants3.