Figure 1. Generation of CISH-KO NK cells from human iPSCs.
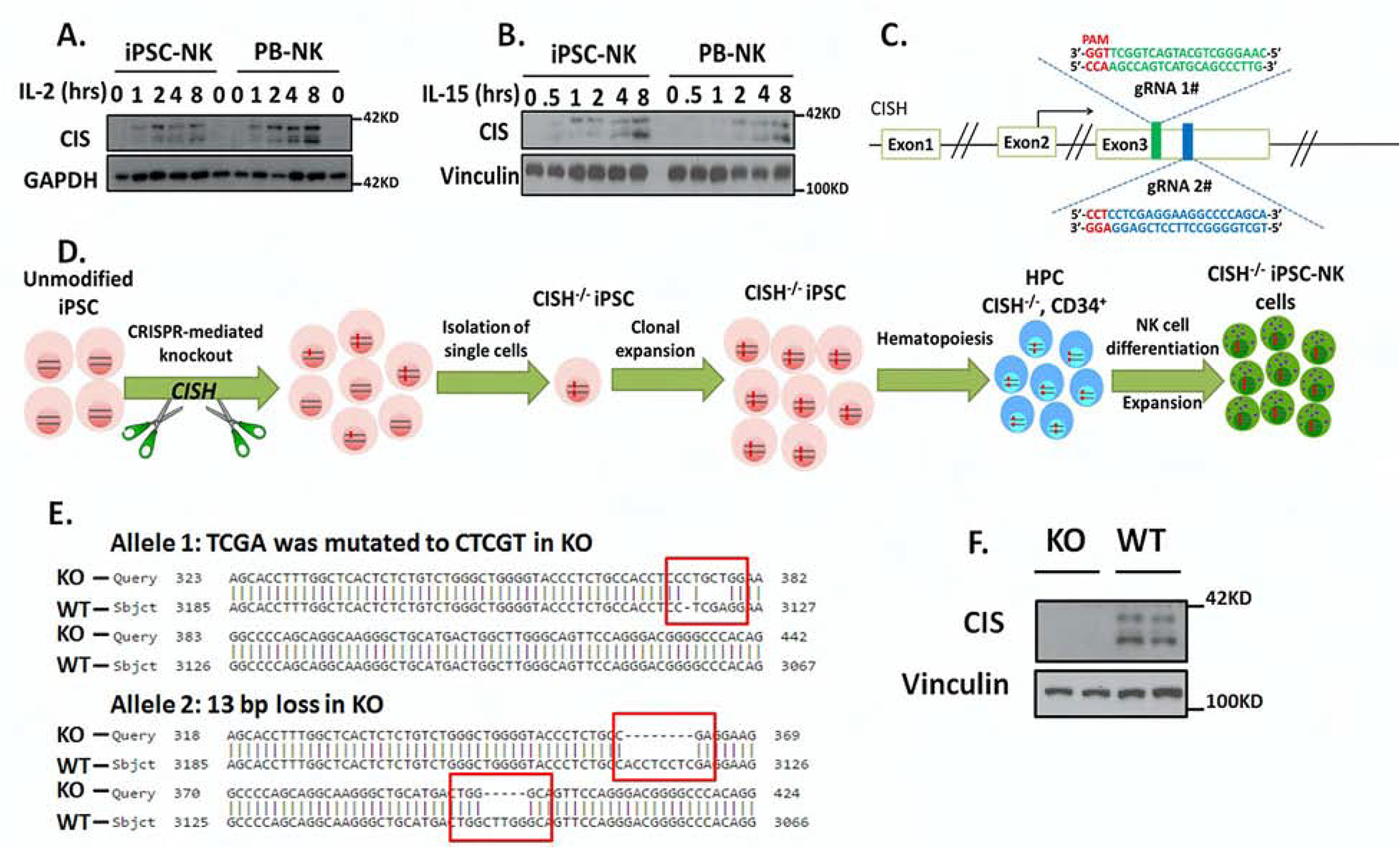
A–B. To demonstrate normal CIS expression iPSC-NK and PB-NK cells were incubated without cytokines for 8hrs and then simulated with (A) 100 U/ml IL-2 or (B) 10ng/ml IL-15 for the indicated times, then CIS expression was analyzed by immunoblotting (IB). GAPDH was used as loading control. C. Scheme of CRISPR/Cas9-mediated CISH KO using two guide RNAs (gRNA) located in direct and complementary strand targeting exon 3 of the CISH gene. D. Schematic representation for deriving clonal CISH−/− iPSC-NK cells from human iPSC. CRISPR/Cas9 mediated CISH KO was performed in WT iPSC, followed by identification of CISH−/− iPSC at clonal level. After clonal expansion, CISH−/− iPSC were differentiated to CD34+ hematopoietic progenitor cells through hematopoiesis and then CISH−/− iPSC-NK cells through NK cell differentiation using the method previously reported. E. Comparison of sequence in CISH KO clone (obtained by Sanger sequencing) with CISH WT sequence (exon3, from 3067 to 3185) by Basic Local Alignment Search Tool (BLAST) showing frame shift mutations (red rectangle) in both alleles. All mutations occurred in 2 gRNA targeted region. F. WT-iPSC-NK cells and CISH−/− iPSC-NK cells were simulated with 10 ng/ml IL-15 for 8 hours and CIS expression evaluated by IB. Vinculin was used as loading control. Data at A, B and F were repeated in 3 separate experiments.
