Figure 1.
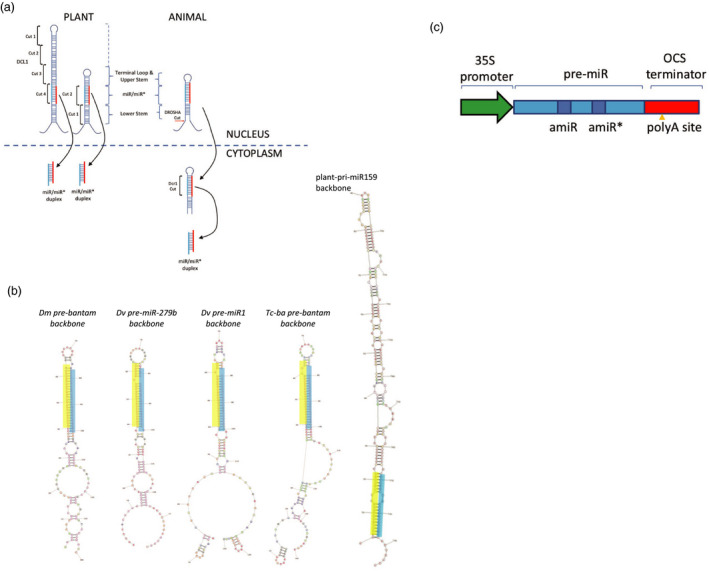
plin‐amiR design and constructs. (a) biogenesis of miRs in plants and animals. Plant pre‐miRs processed in nucleus either by top‐down or bottom‐up cleavage by Dicer‐like 1 (DCL1). Animal pri‐miR cleaved by Drosha in nucleus to form pre‐miR which is exported to cytoplasm where it is processed by Dicer 1 (DCR1). Note longer upper and lower stems in plant precursors than in animal structures. (b) Structures of plin‐amiR sequences targeting the acetylcholinesterase gene of H. armigera. Plant artificial miRNA (At‐miR159) based on miR‐159 and insect backbones: (Dm‐ba) Drosophila melanogaster bantam scaffold, (Dv‐miR279) Diabrotica virgifera virgifera mir‐279b scaffold, (Dv‐mir1) D. v. virgifera miR‐1 scaffold, (Tc‐ba) Tribolium castaneum bantam scaffold. Each insect miRNA backbone contains 40‐nucleotide flanking regions from miRNA primary transcripts listed above. The sequence that is antisense to H. armigera ACE2 is highlighted in cyan. RNAstructure version 5.5 (Mathews lab, http://rna.urmc.rochester.edu/RNAstructure.html) was used to generate the secondary structure. (c). Schematic of plin‐amiR construct.
