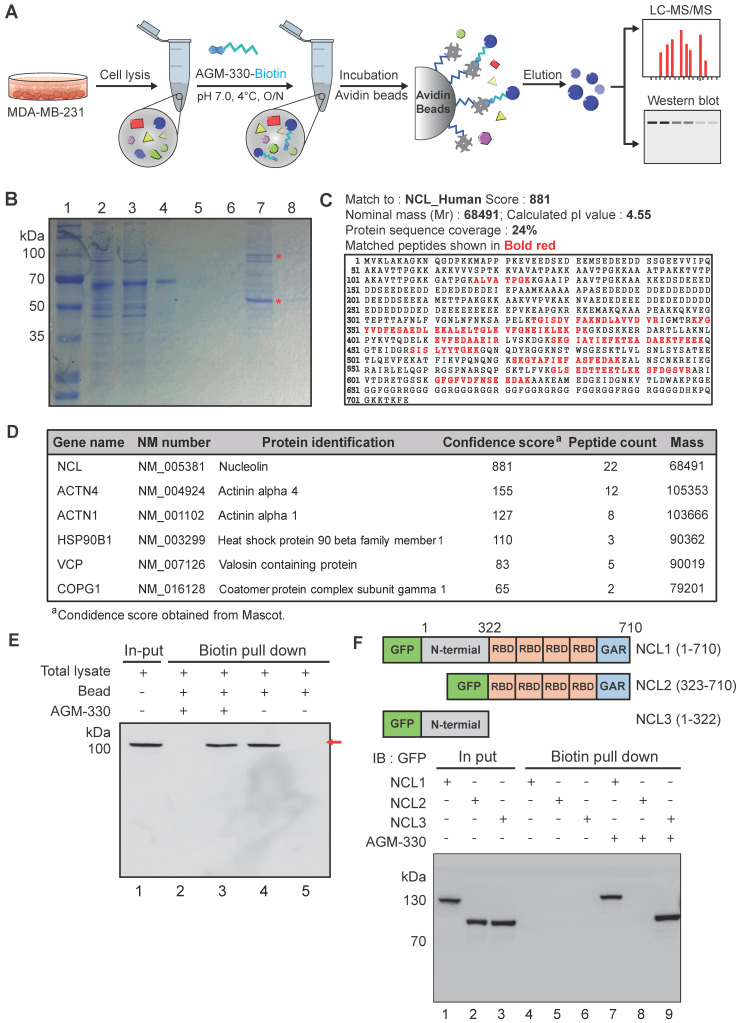
Figure 2.
Characterization and identification of molecular targets of AGM-330. A, Schematic diagram of biotin pull-down assay, mass spectrometry analysis and Western blot. B, Coomassie brilliant blue staining of the eluted proteins from affinity columns after separation by 10% SDS-PAGE. Lane 1: protein marker; Lane 2: total lysate before biotin pull-down assay; Lane 3: flow-through after incubation with AGM-330-biotin; Lanes 4-6: bead washing fraction; Lanes 7-8: elution of AGM-330 binding proteins. * Red star indicates that the protein band was excised for in-gel digestion followed by LC-MS/MS analysis. C, Protein database searches of peptides detected by MS identified NCL as an AGM-330 binding partner. D, List of genes obtained from LC-MS/MS analysis. The confidence score indicates the mascot score of the identified protein. E, Immunoblotting analysis of the eluted proteins from the biotin pull-down assay using an anti-NCL antibody. The arrow indicates the presence of NCL. Lane 1: Input of total lysate; Lane 2: flow through after incubated with AGM-330-biotin; Lane 3: elution of AGM-330 binding proteins; Lane 4: flow through after incubated with beads (no AGM-330); Lane 5: elution of beads binding proteins (no AGM-330). F, Analysis of the NCL domains binding to AGM-330. Different GFP-NCL constructs were transfected into HEK293T cells. AGM-330-biotin was added to NCL residues 1-710, residues 1-322, or residues 323-710 bound to streptavidin beads. Proteins on the beads were analyzed using immunoblotting with anti-GFP antibodies. Lanes 1-3: Input of total lysate; Lanes 4-6: elution after incubation with beads (no AGM-330); Lanes 7-8: elution of AGM-330 binding proteins.