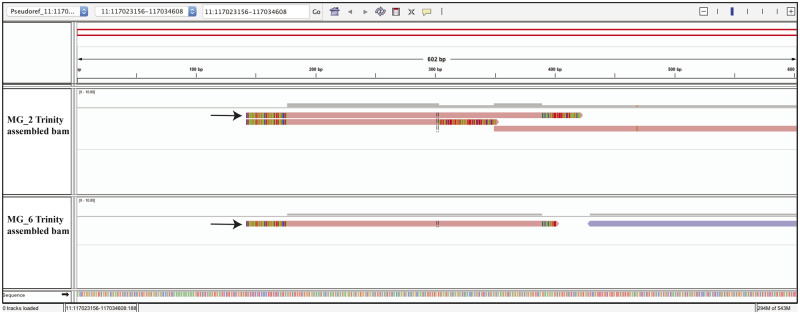
Figure 4:
Integrated Genomics Viewer (IGV) screen shot of a circRNA candidate (chr11:117 023 156–117 034 608) Assembled contigs generated by ACValidator on RNase R-treated (top panel) and nontreated (bottom panel) human MG samples, aligned to the corresponding pseudo-reference. This circRNA was detected in both the treated and nontreated samples by at least three of the six existing circRNA prediction algorithms, and was not depleted following RNase R treatment. The circRNA junction of interest is at 300 bp, and pink bars that span over this junction represent the contigs that validate the junction (colored segments at the ends of contigs represent soft-clipped bases; arrows indicate the generated contigs that overlap with the circRNA junction). Reads from these samples aligned to the reference are shown in Supplementary Figure 3.