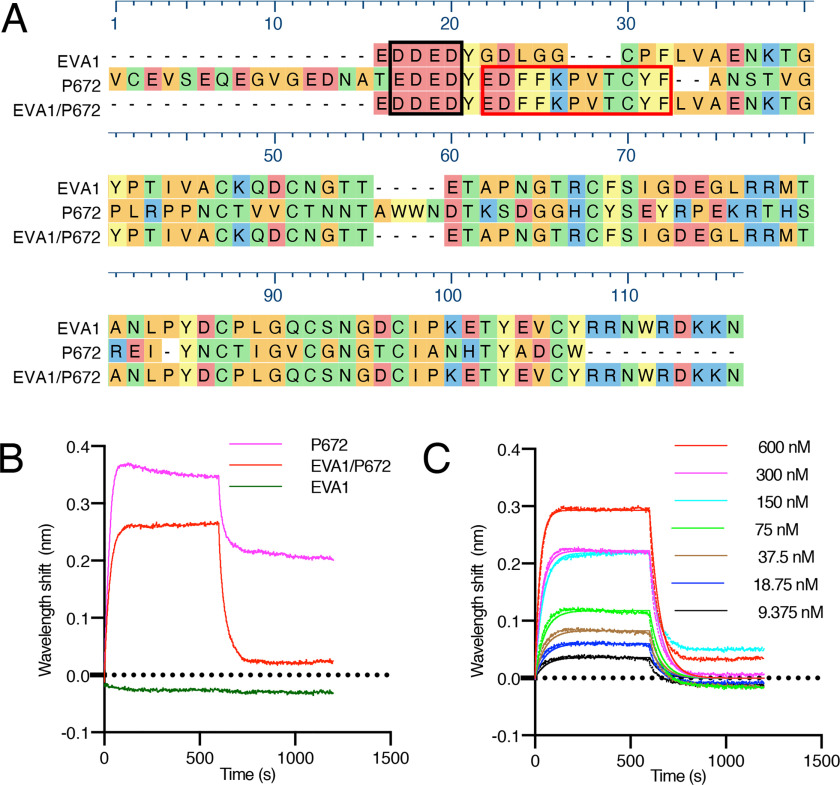
Figure 2.
Design and biophysical analysis of an EVA1/P672 hybrid protein. A, alignment of EVA1, P672, and EVA1/P672 (EVA1 containing P672E22–E32) hybrid protein using MUSCLE algorithm. Amino acids are color-coded according to physicochemical properties: yellow, aromatic (Phe, Trp, and Try); red, acidic (Asp and Glu); blue, basic (Arg, His, and Lys); orange, nonpolar aliphatic (Ala, Gly, Ile, Leu, Met, Pro, and Val); and green, polar neutral (Cys, Asn, Gln, and Thr). Amino acids that were protected from deuterium uptake in P672 are indicated with a red box. The N-terminal acidic region is enclosed in a black box. B, biolayer interferometry sensorgram obtained when either P672, EVA1/P672, or EVA1 is loaded onto the biolayer interferometry sensor and exposed to 600 nm CCL8. Plots display wavelength shift (y axis, nm) versus time (x axis, s). C, biolayer interferometry sensorgram for EVA1/P672 hybrid binding to CCL8. Dotted lines indicate collected data, and solid lines indicate modeled data. Plots display wavelength shift (y axis, nm) versus time (x axis, s).