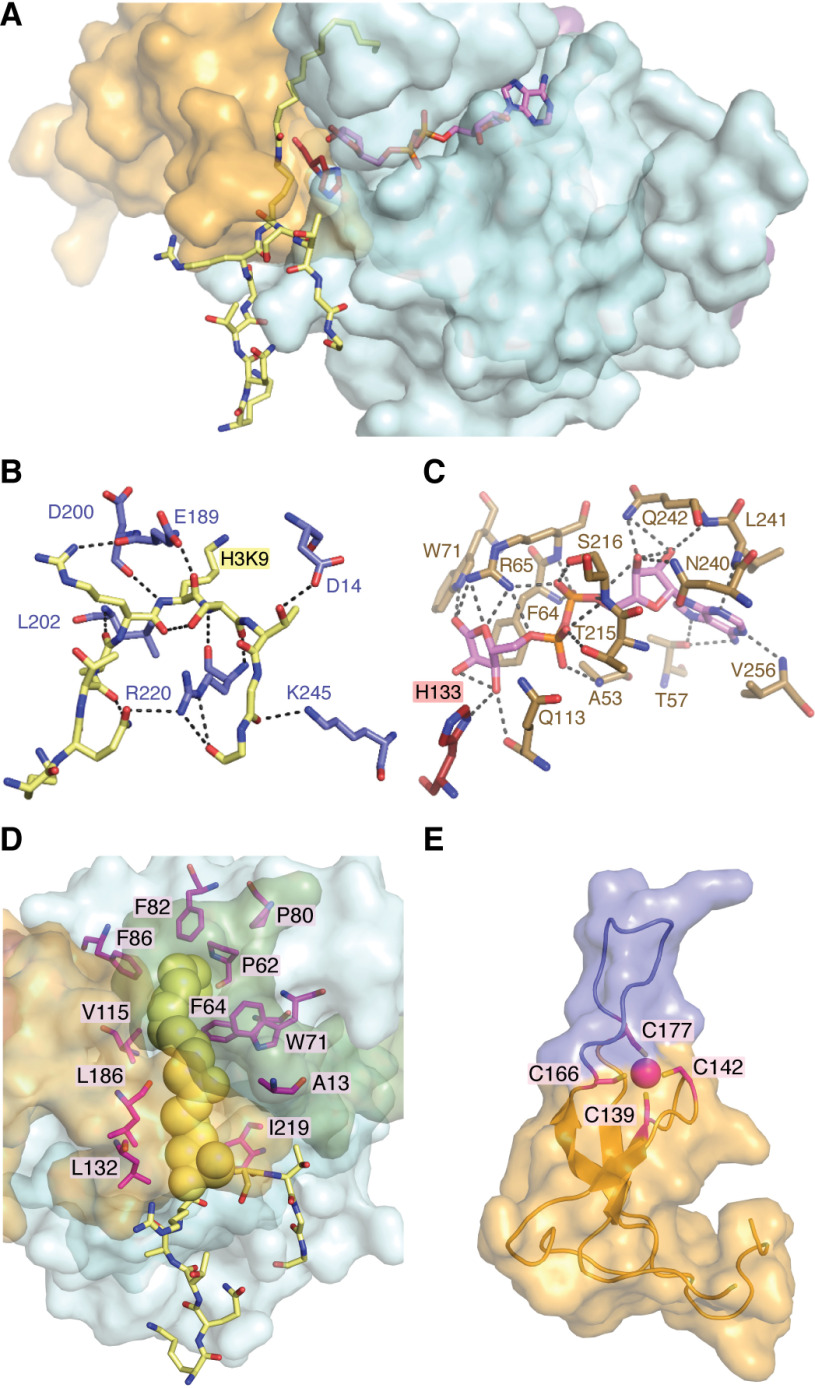
Figure 4.
SIRT6 structural features of ligand binding and overall architecture. SIRT6 sequentially binds NAD+ followed by acyl-substrate prior to initiating chemical catalysis. The noncatalytic NAD+ surrogate lacking nicotinamide, ADP-ribose, is often used in structural studies to prevent catalytic turnover. A, ADP-ribose (pink) binds to the interior of the Rossman fold domain with the nicotinamide-ribose proximal to the catalytic His-133 (red). H3K9 myristoyl peptide (yellow) binds to the solvent-exposed exterior of both the Rossmann fold and Zn2+-binding domain with the substrate lysine inserted into the catalytic site proximal to the opposing side of His-133. The N terminus has been excluded to more clearly demonstrate the relative positioning of ligands. B, polar contacts between the H3K9 peptide substrate (yellow) and residues SIRT6 (light blue). C, extensive polar contacts between residues of SIRT6 (tan) and ADP-ribose (pink). D, SIRT6 contains an extended hydrophobic tunnel, relative to the latent tunnels observed in other structurally solved sirtuins, that accommodates the myristoyl chain (yellow spheres) of an H3K9myr peptide (yellow sticks). This hydrophobic tunnel is formed between the Rossmann fold domain (light blue surface) and Zn2+-binding domain (orange surface), is composed of numerous nonpolar residues (magenta sticks), and is sealed by the N terminus (green surface). E, the Zn2+-binding domain of SIRT6 utilizes four conserved cysteines (magenta sticks) to coordinate a zinc-ion (magenta sphere). This domain contains a 10-amino acid extension between the third and fourth cysteines, resulting in an unordered loop (blue) unique to SIRT6. These structures were produced using entry 3ZG6 from the RCSB Protein Data Bank.