Figure 1. Meta-analysis of existing datasets corroborated major neurodegenerative disease hypotheses but revealed gaps in understanding how neural cell heterogeneity influences AD-associated dysfunctional mechanisms.
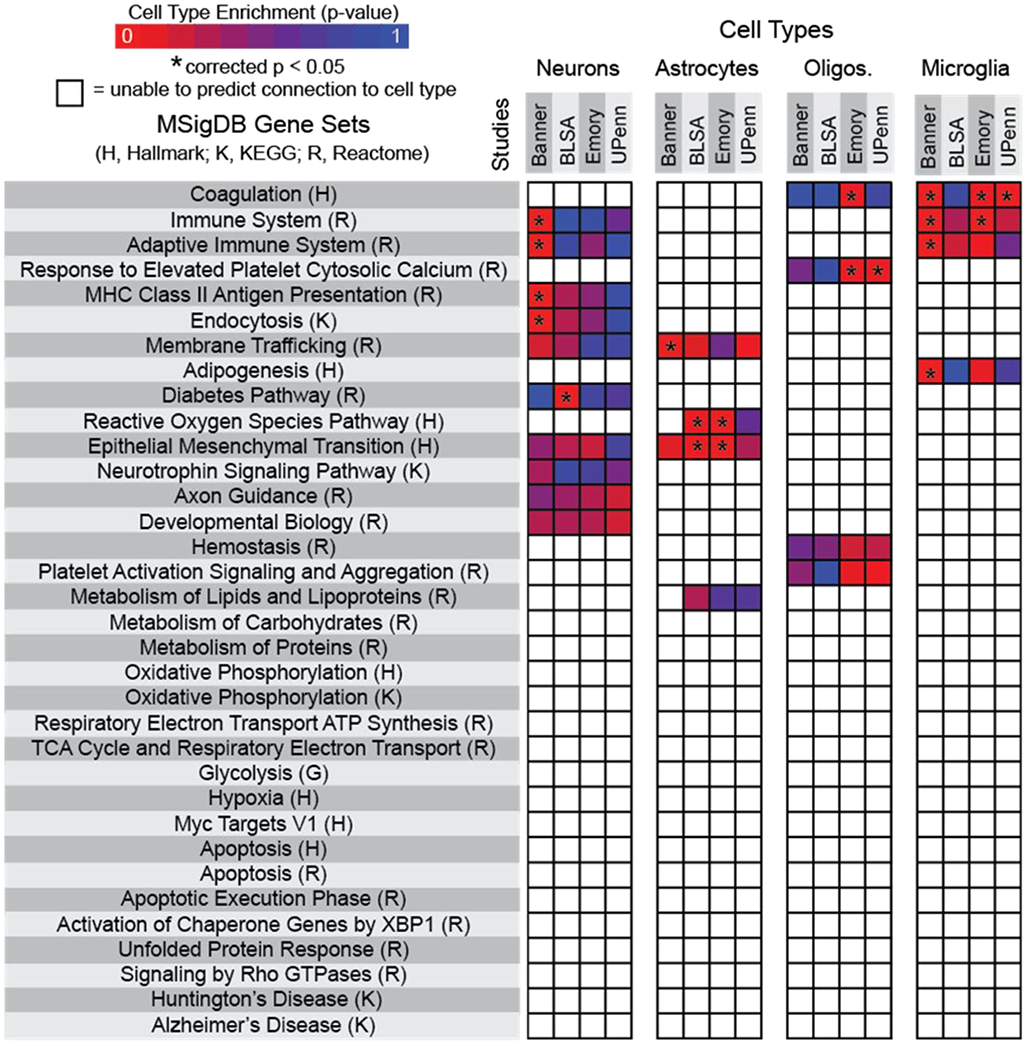
Four datasets (Banner Sun Health Research Institute Study (Banner), Baltimore Longitudinal Study on Aging Study (BLSA), Emory University Alzheimer’s Disease Research Center Brain Bank Study (Emory), and the Perelman School of Medicine University of Pennsylvania Proteomics Pilot Study (UPenn)) were assessed1,16,37,41. Listed gene sets include all pathways significantly different in Alzheimer’s disease versus non-symptomatic proteomic profiles. Heat maps represent specific cell type predictive contributions to dysfunctional pathways, with asterisks indicating statistical significance (corrected p < 0.05). Non-colored boxes indicate inconclusive tests in which a p-value was unable to be generated.
