Figure 2. After processing, fresh and paraformaldehyde (PFA)-fixed rat brain cells exhibit similar numbers, amounts, and cell component localization of proteins.
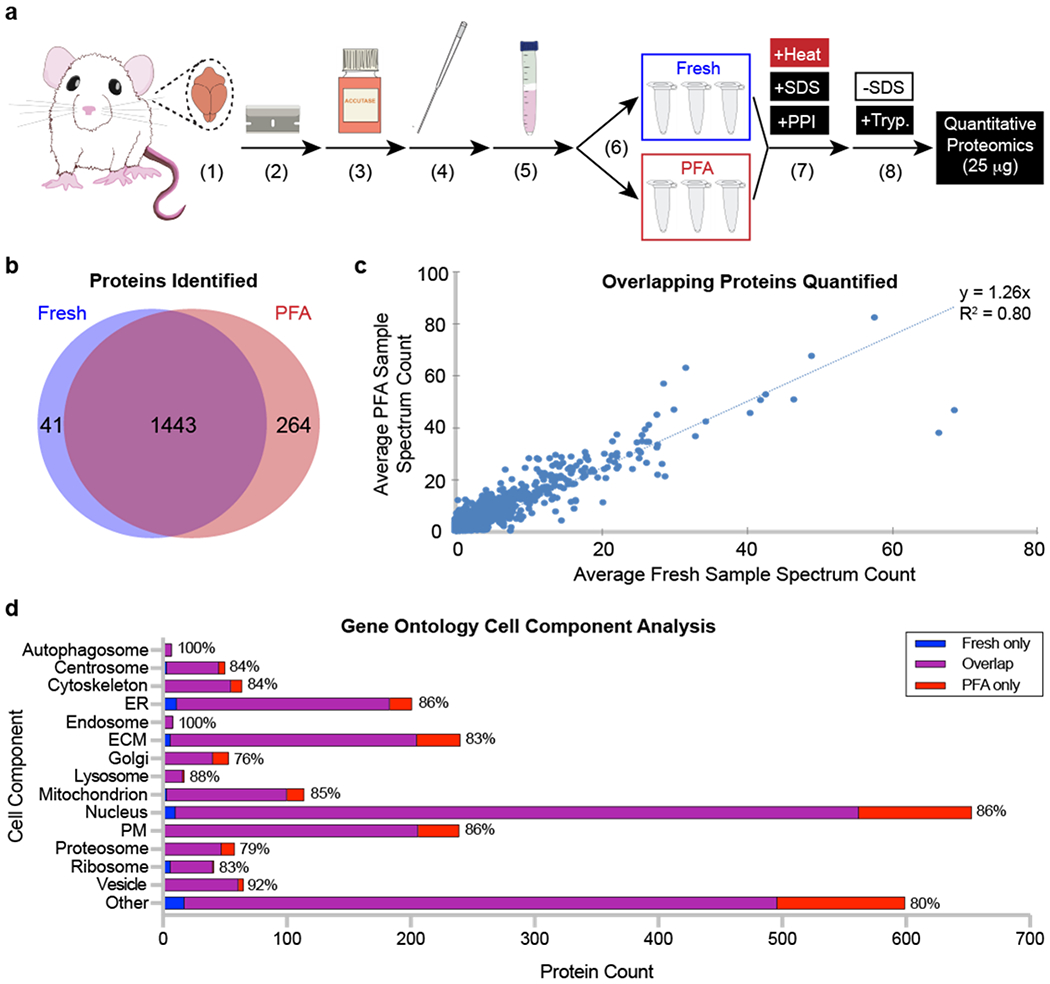
(a) Rat sample processing. A rat brain was (1) isolated, (2) diced into small pieces, and dissociated using (3) enzymatic digestion and (4) mechanical trituration. (5) Cells were isolated using a Percoll gradient before (6) being split into fresh and PFA samples (N = 3 per condition). (7) All samples were processed using detergent and high temperature. (8) Sodium dodecyl sulfate (SDS) was removed from protein lysates, and samples were digested using trypsin before being analyzed by tandem mass spectrometry. (b) Unique proteins identified in fresh and PFA samples. (c) Quantity of unique proteins identified compared between fresh and PFA samples. (d) Cell component localization from fresh only, overlapping, and PFA only identified proteins. Percentages at the end of each bar represent the proportion of overlapping proteins compared to the total number of proteins identified across all conditions. Abbreviations: ER, endoplasmic reticulum; ECM, extracellular matrix; PM, plasma membrane; PPI, protease and phosphatase inhibitor; SDS, sodium dodecyl sulfate; Tryp., trypsin.
