Fig. 2. Characterizing TF modules driving concurrent, dynamic gene regulatory processes in populations of single cells.
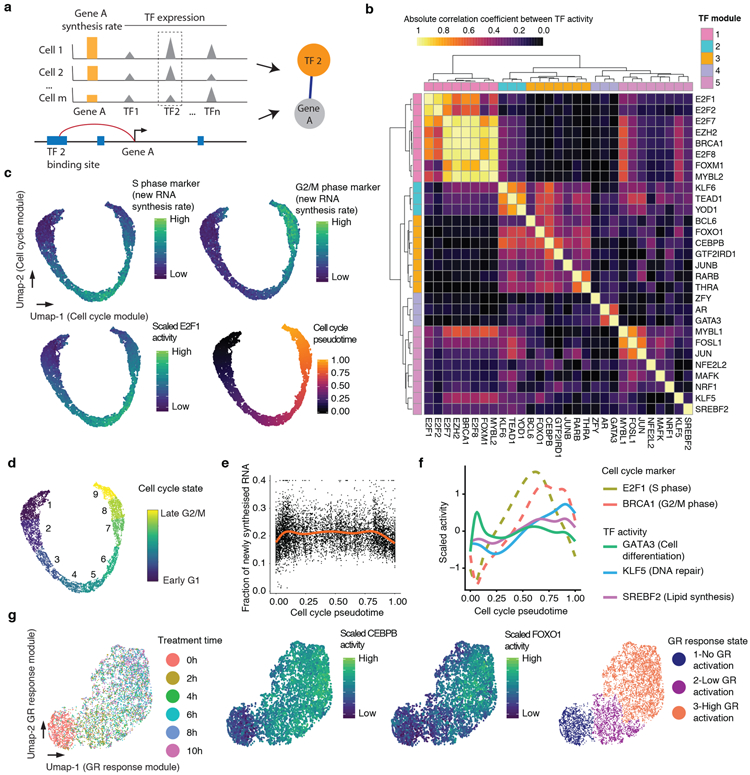
(a) Schematic of approach used to identify links between TFs and their regulated genes. (b) Heatmap showing the absolute Pearson’s correlation coefficient between the activities of pairs of TFs (Cell number n = 6,680). (c) UMAP visualization of A549 cells (n = 6,680) based on the activity of cell cycle-related TF module, colored by levels of newly synthesized mRNA corresponding to S phase markers (top left), G2/M phase markers (top right), and E2F1 activity (bottom left). Bottom right panel is colored by pseudotime based on point position on the principal curve estimated by princurve package65. (d) Same as panel c, but colored according to nine cell cycle states defined by unsupervised clustering analysis. In broad terms, cell cycle states 1-3 correspond to G1 phase, 4-6 to S phase, and 7-9 to G2/M phase. (e) Scatter plot showing the changes in the fraction of newly synthesized mRNA in each cell (n = 6,680) along cell cycle progression. The red line is the smoothed curve estimated by the geom_smooth function66. (f) Similar to panel e, but showing smoothed activity of selected TF modules as a function of cell cycle pseudotime. (g) UMAP visualization of A549 cells (n = 6,680) based on the activity of GR response-related TF module, colored by DEX treatment time (left), CEBPB or FOXO1 activity (middle panels), or cluster id from unsupervised clustering (right). Throughout figure, to calculate TF module activity, newly synthesized UMI counts for genes linked to module-assigned TFs are scaled by library size, log-transformed, aggregated and then mapped to Z-scores.
