Figure 3. Mutational analysis and N-terminal truncation of SPAM shows the residues critical for hPD-L1 binding.
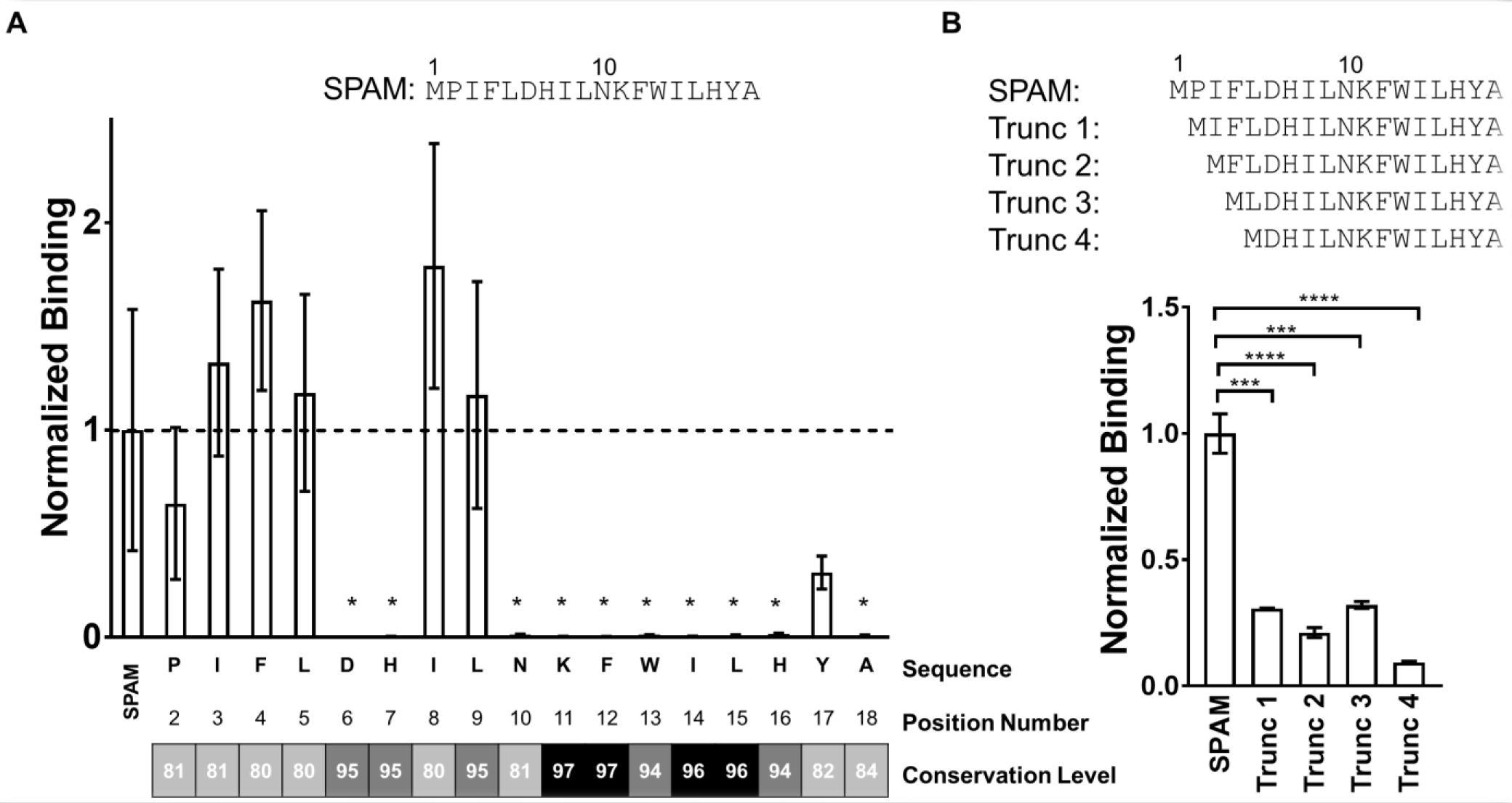
A) Mutational analysis of SPAM; [35S]-labeled SPAM variants were translated with alanine substituted at each position (2–18) and glycine substituted for alanine at position 18. hPD-L1 binding of each [35S]-SPAM alanine mutant was measured and normalized to the binding of [35S]-SPAM. The mean of the normalized binding for each SPAM variant (n=3) is shown along with the standard deviation of the mean. The conservation level for each doped residue in X17 library was obtained from next-generation sequencing data. These results were binned according to sequence family and 108 sequences from SPAM family that appeared more than 1,000 times were identified and the percent conservation relative to the SPAM peptide calculated for each position. B) [35S]-labeled N-terminally truncated versions of SPAM were translated where one (P), two (P, I), three (P, I, F) and four (P, I, F, L) residues were removed from the N-terminus of SPAM creating Trunc 1, Trunc 2, Trunc 3 and Trunc 4, respectively. hPD-L1 binding of each truncated variant was measured and normalized to the binding of [35S]-labeled SPAM. Each measurement was carried out in triplicate and error bars show the standard deviation of the mean normalized binding. (Students t-test; p value < 0.05 (*), < 0.01 (**), < 0.005 (***), < 0.001 (****)).
