Summary
Microglia are the innate immune cells of the central nervous system. Although numerous methods have been developed to isolate microglia from the brain, the method of dissociation and isolation can have a profound effect on the function of these highly dynamic cells. Here, we present an optimized protocol to isolate CD11b+ cells (microglia) from mouse or human brain tissue using magnetic bead columns. Isolated microglia can be used to model diseases with neuroinflammatory components for potential therapeutic discoveries.
For complete details on the use and execution of this protocol, please refer to Hanamsagar et al., 2017, Rivera et al., 2019, and Edlow et al. (2019)
Graphical Abstract
Highlights
-
•
An optimized protocol for isolation of mouse or human microglia
-
•
Use of magnetic beads for CD11b+ cell isolation
-
•
Adaptations for isolation from embryonic or adult tissue
-
•
Adaptable for different endpoints (e.g., RNA, protein, flow cytometry, cell culture)
Microglia are the innate immune cells of the central nervous system. Although numerous methods have been developed to isolate microglia from the brain, the method of dissociation and isolation can have a profound effect on the function of these highly dynamic cells. Here, we present an optimized protocol to isolate CD11b+ cells (microglia) from mouse or human brain tissue using magnetic bead columns. Isolated microglia can be used to model diseases with neuroinflammatory components for potential therapeutic discoveries.
BEFORE YOU BEGIN
Can Be Made Prior to Day of Procedure
Prepare the MACS Buffer before Starting the Procedure
(Refer to Materials and Equipment for buffer recipe).
Preparation of Nylon Filters
Cut nylon Nitex mesh fabric rolls (Sefar #03-70/33) into approximately 3cm x 3cm squares, placed into a beaker, and autoclaved prior to placement in cell culture hood.
Alternatives: While our lab uses Sefar nylon mesh filter rolls, any alternative 70 μm filter will suffice (e.g., Miltenyi MACS SmartStrainers).
Preparation of #1, #2, #3 Pasteur Pipets
Successively smaller Pasteur pipets are created by flame polishing FisherScientific Fisherbrand™ Disposable Cotton-Plugged Borosilicate-Glass Pasteur Pipets. The largest opening pipet is referred to as #1, the second largest as #2, and the smallest as #3. Due to the nature of flame-polishing pipets, every individual pipet will have some variation, however we have determined that on average #1 pipets have an opening of 0.75–0.8 cm cm, #2 have an opening of 0.5–0.55 cm, and #3 have an opening of 0.2–0.25 cm (Video S1, Figure 1).
Figure 1.
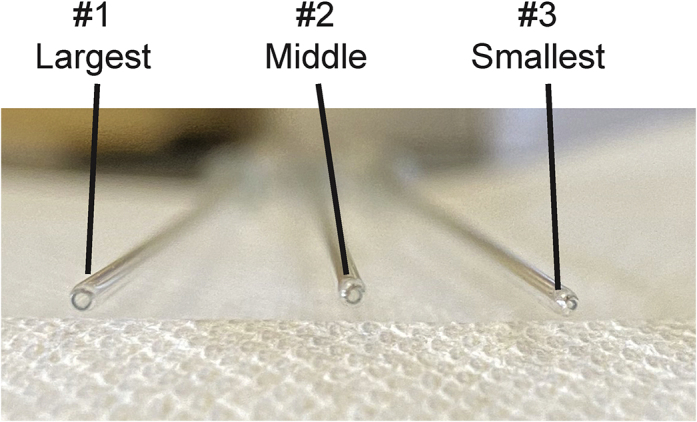
Representative Images for #1, #2, and #3 Pasteur Pipets
While our lab dissociates tissue by shear force of successively smaller Pasteur pipets, other dissociation methods may prove acceptable. For instance, the Miltenyi gentleMACS Dissociator (Miltenyi #130-093-235) can be used to dissociate tissue prior to subsequent steps.
While the method of force used to dissociate tissues may vary, we do recommend heat and enzyme-mediated digestion be used for this protocol (See Limitations)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| HBSS (without calcium/magnesium) | ThermoFisher Scientific | 14175-095 |
| HBSS (with calcium/magnesium) | ThermoFisher Scientific | 14025-092 |
| HyClone Characterized Fetal Bovine Serum, U.S. Origin | GE Healthcare | SH30071.03 |
| HEPES (1 mM) | ThermoFisher Scientific | 15630-080 |
| Collagenase A | Millipore Sigma | 11088793001 |
| DNase I, grade II, from bovine pancreas | Millipore Sigma | 10104159001 |
| Percoll | Millipore Sigma | GE17-0891-01 |
| Debris Removal Solution | Miltenyi Biotec | 130-109-398 |
| PBS (1X), pH 7.4 | ThermoFisher Scientific | 10010-023 |
| PBS (10X), pH 7.4 | ThermoFisher Scientific | 70011-044 |
| EDTA (0.5 M), pH 8.0, RNase-free | ThermoFisher Scientific | AM9261 |
| Bovine Serum Albumin | Millipore Sigma | A9647 |
| Sodium Pyruvate (100 mM) | ThermoFisher Scientific | 11360-070 |
| CD11b (Microglia) Microbeads, human and mouse | Miltenyi Biotec | 130-093-634 |
| Experimental Models: Organisms/Strains | ||
| C57BL6J Mice | The Jackson Laboratory | 000664 |
| Postmortem Human Motor Cortex | n/a | n/a |
| Other | ||
| MACS Separation Columns (LS) | Miltenyi | 130-042-401 |
| QuadroMACS Separator | Miltenyi | 130-090-976 |
| MACS MultiStand | Miltenyi | 130-042-303 |
| Refrigerated Centrifuge | Eppendorf | 5810R |
| Rotor with Buckets | Eppendorf | A-4-81 |
| Nitex Open Mesh Fabric | Sefar | 03-70/33 |
| Cotton-Plugged Borosilicate-Glass Pasteur Pipet | FisherScientific | 13-678-8B |
MATERIALS AND EQUIPMENT
Alternatives: While we use Sefar nylon filters that are cut and sterilized, commonly used alternatives such as Miltenyi 70 μm filter should be equivalent.
While we use an Eppendorf 5810R refrigerated centrifuge with A-4-81 rotor with swinging buckets, any refrigerated centrifuge with swinging buckets and an ability to finely control brake and acceleration speeds should suffice. Acceleration and deceleration settings should be validated for alternative equipment
While we use Miltenyi MACS Separation Columns with Miltenyi MACS MultiStand magnet, other positive selection columns and magnets (such as BioLegend MojoSort Magnet and R&D Systems MagCellect Magnet) may be an acceptable alternative. Validation must be performed if using equipment from alternative suppliers.
Note: Validation must be performed if reagents are used from alternative suppliers.
MACS Buffer
Preparation of MACS Buffer
First, dissolve EDTA and BSA in 500 mL of 1X PBS. Then raise volume to 1 L with 1X PBS. Vacuum filter slowly for 30 min in tissue culture hood. Keep sterile.
| Solution | Amount |
|---|---|
| 1X PBS | 1 L |
| EDTA | 1 mM |
| Bovine Serum Albumin | 1% |
To Be Made Fresh Day of Procedure
Note: If performing the Percoll adaptation of this protocol, Percoll, 1X PBS, and 10X PBS should be taken out of the refrigerator and placed at ~20°C–25°C prior to beginning the rest of the protocol.
Note: All procedures following cardiac perfusion with sterile saline and tissue dissection should be performed in a Class II biological hood with standard aseptic technique.
Enzyme Digestion Mix
Preparation of Enzyme Digestion Mix
Note: Collagenase A powder can be challenging to dissolve in the enzyme digestion mix.
| Solution | Amount |
|---|---|
| HBSS (without calcium/magnesium) | |
| Fetal Bovine Serum | 5% |
| HEPES (1 mM stock) | 10 μM |
| Collagenase A | 2 mg/mL |
| DNase I | 28 U/mL |
Isotonic Percoll
Preparation of Isotonic Percoll
CRITICAL: All elements for the Percoll gradients should warm to ~20°C–25°C prior to mixing
| Solution | Amount |
|---|---|
| PBS (10X) | 10% |
| Percoll | 90% |
70% Percoll Solution
4 mL of 70% Percoll is required per sample
CRITICAL: All elements for the Percoll gradients should come to ~20°C–25°C prior to mixing
| Solution | Amount |
|---|---|
| PBS (1X) | 30% |
| Isotonic Percoll | 70% |
30% Percoll Solution
6 mL of 30% Percoll is required per sample
CRITICAL: All elements for the Percoll gradients should come to ~20°C–25°C prior to mixing
| Solution | Amount |
|---|---|
| PBS (1X) | 70% |
| Isotonic Percoll | 30% |
Post-Percoll Solution
10 mL of Post-Percoll Solution is required per sample
| Solution | Amount |
|---|---|
| HBSS (with calcium/magnesium) | |
| Fetal Bovine Serum | 5% |
| HEPES | 10 μM |
STEP-BY-STEP METHOD DETAILS
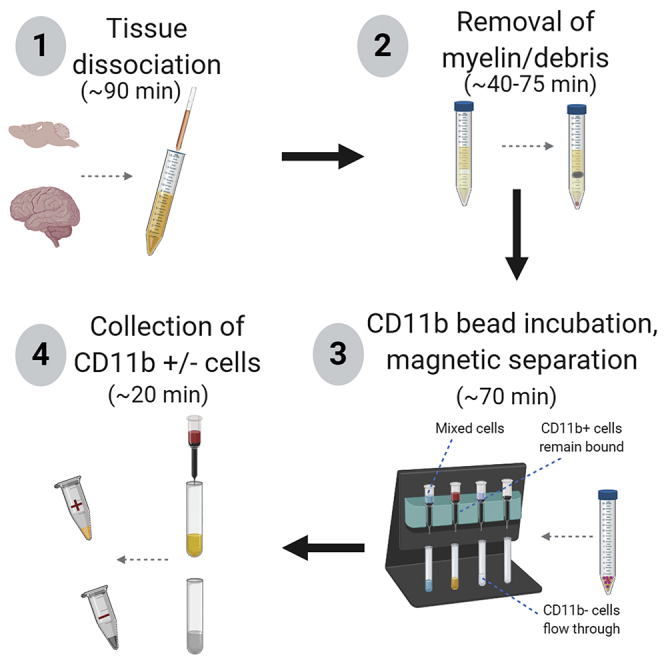
Timing for Steps 1–7: (~90 min from end of transcardiac perfusions)
This protocol can be adapted depending upon the type of input material as well as the endpoint assay (Figure 2). The protocol can be shortened for time-sensitive endpoints such as RNA extraction. The protocol can also use Percoll or Debris Removal Solution to remove myelinated material and other debris for assays in which cells will be cultured, and to isolate microglia in the interphase layer (Percoll). While our lab more commonly uses the Debris Removal Solution protocol when isolating microglia from adult mouse brain tissue, we use the Percoll method for embryonic mouse tissue, as the Debris Removal Solution did not successfully pellet any cells from embryonic tissue (see Limitations).
-
1.
Aliquot 5 mL of Enzyme Digestion Mix into 15 mL tubes and weigh tubes.
-
2.
Saline transcardiac perfusion (see Note), dissect brain region of interest, chop up with razor blades on ice, and place in cold enzyme digestion mix on ice - for human tissue chop up with razor blades and place in cold enzyme digestion mix on ice.
Note: Not for embryonic tissue
Note: We have tested this protocol using human postmortem fresh frozen motor cortices provided by the Massachusetts Alzheimer’s Disease Research Center (ADRC) with approval from Massachusetts General Hospital IRB (1999p009556). Postmortem tissue was obtained as follows: briefly, the brain hemisphere was removed, coronally sectioned, and fresh frozen on metal plates, after which tissue was stored in a -80°C freezer until Step 2.
Note: We have performed this protocol from mouse tissue using whole brain or dissected regions such as the prefrontal cortex, hippocampus, and nucleus accumbens. While we have not tested this protocol from every region of the mouse brain, we believe that it is capable of isolating CD11b+ cells from all regions. Specific regions should be optimized for use.
Note: Following transcardiac perfusion of mouse with ice cold saline, it is necessary to dissect brain regions of interest/chop of tissue on an ice-cold petri dish using ice-cold dissection tools.
-
3.
Weigh samples (make sure to wipe water off the side of tube with kim wipe). Note the weight of the sample (Measurement in Step 3 vs measurement in Step 1), as this input will influence the volume of beads and buffer added in Step 23.
Note: Make sure to wipe water off the side of tube prior to weighing
-
4.
Place samples in a 37°C water bath for 15 min.
CRITICAL: Agitate every 5 min. Agitate tubes, but do not invert to mix, as tissue has the tendency to become stuck near the lid of 15 mL tubes.
-
5.
Dissociation: Dissociate tissue to a single-cell suspension over the course of three separate flame-polished Pasteur pipets (first use #1, then #2, then #3). Pipet up and down 20 times with each pipet. Incubate at 37°C for 15 min in between each numbered pipet step. (Video S2, Figure 3)
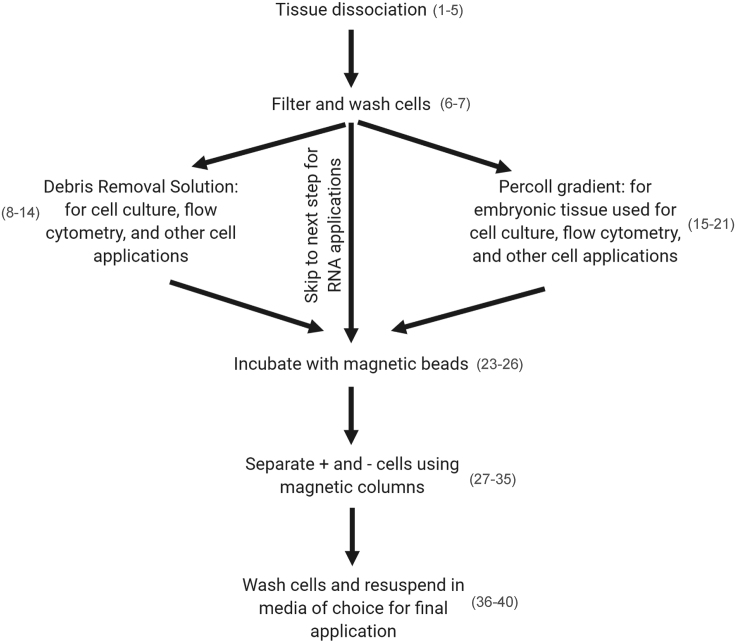
Figure 2.
Workflow of Isolation Protocol with Corresponding Steps in Parentheses
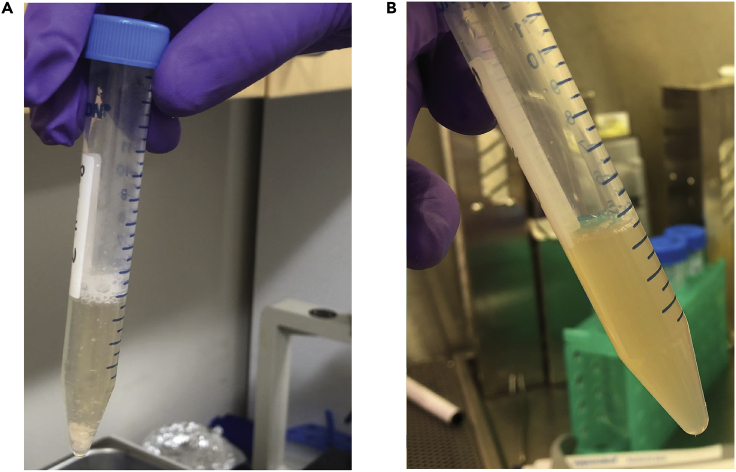
Figure 3.
Dissociation of Tissue to a Single-Cell Suspension
Appearance of tissue (A) prior to and (B) after dissociation steps
CRITICAL: Agitate every 5 min. For instance, pipet up and down 20 times using #1 pipet, then agitate tubes every 5 min during the 15 min incubation step. Then begin pipetting with #2 pipet.
Pipet aggregated tissue until all tissue moves freely up into the Pasteur pipet. Then pipet up and down 20 times.
During agitation steps, prepare the 30% and 70% percoll solutions (that were previously brought to ~20°C–25°C if using percoll gradients/embryonic tissue.
-
6.
Filter samples into fresh 15 mL tubes. Place 70 μm nylon filters on top of fresh 15 mL tube. Pre-wet each filter as below (Video S3). Add cell suspension over the pre-wet area on each filter. Rinse old 15 mL tubes with 2 mL of HBSS (with calcium/magnesium. Pipet up and down to mix. Add over the filter. Fill the new tubes to 15 mL with HBSS (with calcium/magnesium).
CRITICAL: Pre-wet each filter by adding 1 mL of HBSS (with calcium/magnesium) to the middle of the filter and then drag to side.
CRITICAL: After this step, samples should be kept on ice as much as possible, except in the case of the Percoll gradients, which will be at ~20°C–25°C again after Step 7, in preparation for the 23°C spin.
-
7.
Centrifuge: 10 min, 300 × g, 4°C, max acceleration, half brake.
Note: At this point, the protocols diverge depending upon input tissue or endpoint methods. Choose one of the methods below to continue the protocol at step 8, step 15 or step 22.
Debris Removal Solution Protocol
Timing for Steps 8–14: (~30–40 min)
For general use for cell applications. This section will remove debris/myelin from single-cell suspension prior to isolation of CD11b+ cells.
-
8.
Aspirate the supernatant, then resuspend cell suspension carefully with 3,100 μL of ice-cold 1X PBS and transfer cell suspension to a fresh 15 mL tube. DO NOT VORTEX.
-
9.
Add 900 μL of cold Debris Removal Solution.
-
10.
Mix well by pipetting 10 times slowly up and down.
-
11.
SLOWLY layer 4 mL of 1X PBS on top, creating a visible gradient (Video S4)
-
12.
Centrifuge: 10 min, max speed (at least 3,000 × g), 4°C, max acceleration, max brake (Figure 4)
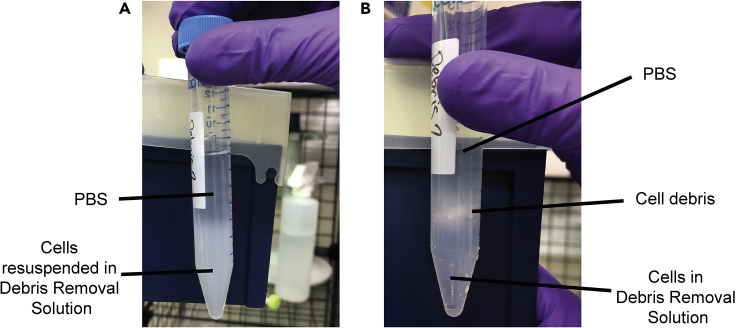
Figure 4.
Debris Removal Solution Gradients
Photographs of example gradients (A) before and (B) after centrifugation.
Begin with tube tilted at 45° angle and slowly layer 1X PBS on top of Debris REMOVAL Solution layer. As more 1X PBS is added, slowly bring tube back to a vertical position.
-
13.
Remove the top two layers. Fill up to 14 mL with ice cold 1X PBS.
-
14.
Centrifuge: 10 min, 1,000 × g, 4°C, max acceleration, max brake.
(To be continued at Step 23)
Percoll Gradient Protocol
Timing for Steps 15–21: (~60–75 min)
For general use for cell applications, especially those utilizing embryonic tissues. This section will remove myelin from single-cell suspension prior to isolation of CD11b+ cells.
Note: Fast temp the centrifuge to 23°C as soon as samples from step 7 are removed.
Note: Ensure that 70% Percoll and 30% Percoll solutions are prepared prior to this step.
-
15.
Aspirate the supernatant, then resuspend cell suspension carefully with 1 mL of 30% Percoll. Then add 4 mL of 30% Percoll on top.
-
16.
Place an unflamed Pasteur pipet in each 15 mL tube (cover the top with a gloved finger while gently placing the tip down to the bottom) and underlay 4 mL of 70% Percoll by pipetting slowly in 1 mL increments into the open top of the Pasteur pipet (Video S5)
CRITICAL: When removing the Pasteur pipet, lift tip very slightly off the bottom of the tube to release any last fluid, and then cover the top of pipet with finger and slowly pull up, taking care not to disturb the layers. See Troubleshooting for what to do if the 70% Percoll won’t drain through the tip of the Pasteur pipet.
Note: There is no visible myelin layer in tissue from embryonic mouse brains
Note: The addition of a colored powder (such as phenol red) to the Isotonic Percoll solution will result in a darker 70% Percoll solution compared to 30% Percoll. This could allow for easier visualization of the gradient. However, the effects of any powder added to the cells would have to be taken into account.
-
17.
Centrifuge: 20 min, 340 × g, 23°C, acceleration of 2, no brake.
CRITICAL: Note the change in temperature, acceleration, and brake from previous steps.
Note: Given the lack of brake, this step takes approximately 28–30 min.
Note: Remove tubes from centrifuge very gently so as not to disturb the gradients.
Note: Fast temp the centrifuge to 4°C after this step.
-
18.
Bring all tubes back to the cell culture hood at ~20°C–25°C. Aspirate out the top 1 mL of liquid. With adult tissues, this would be the myelin layer- due to the relative lack of myelination in the fetal brain, you will either see nothing in the top 1 mL of fluid (30% percoll) or may see a few bits of debris (Figure 5).
Figure 5.
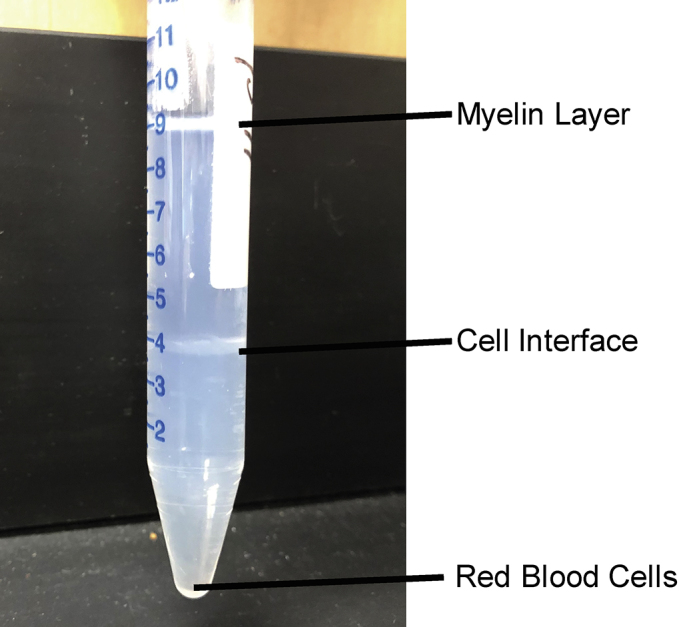
Post-centrifugation Percoll Gradients
Note that myelin will separate at the top of the liquid, cells will separate out at the interface of 70% Percoll/30% Percoll, and red blood cells will pellet at the bottom of the conical tube.
Use finger to cover Pasteur pipet as it is placed under the 30% Percoll. Slowly pipet 70% Percoll into the Pasteur pipet and let it release under the 30% Percoll layer. Cover pipet with finger and gently remove.
-
19.
Pipette out the interphase cells in 1 mL increments four times (total of 4 mLs) and transfer just this 4 mLs containing the cell layer into a new set of labeled 15 mL tubes. Use a circular pipetting method and depress the top of the pipette as you lower the tip down through the 30% percoll to the interphase layer, so you don’t pick up unwanted 30% percoll. See Troubleshooting for tips on visualizing the interphase layer and how to avoid the 70% percoll layer.
-
20.
Fill 15 mL tube up to 15 mL volume with Post-Percoll Solution
-
21.
Centrifuge: 10 min, 300 × g, 4°C, max acceleration, max brake.
CRITICAL: After this step, samples should be kept on ice as much as possible
(To be continued at Step 23)
Molecular Protocol - for time-sensitive molecular applications such as RNA/DNA extraction
-
22.
For this protocol, can move from the centrifugation (Step 7) straight to Step 23 for adult tissues.
For embryonic mouse tissues, steps 15–21 are still required.
Note: With older animals, we have experienced that not removing debris/myelin with Percoll/Debris Removal Solution can clog the column. If column clogging occurs, either of the first two protocols should be used.
∗∗∗All Protocols Converge again at This Step∗∗∗
Timing for steps 23–40: (~90 min)
-
23.
Aspirate the supernatant, then re-suspend pellets with 90 μL MACS buffer and 10 μL CD11b microbeads
CRITICAL: If using a large amount of tissue (i.e. greater than 150 mg), you can increase the amount of MACS buffer/beads. i.e. 135 μL MACS Buffer + 15 μL beads, or 180 μL MACS Buffer + 20 μL beads
Note: 90 μL MACS buffer and 10 μL CD11b microbeads are used when specific brain regions (i.e. hippocampus, nucleus accumbens) of young animals (i.e. PN30-75) are dissected for isolation. Older animals or larger input tissue (i.e. >150 mg) would require more MACS Buffer/beads. We recommend a titration to determine the optimal bead/buffer volume for your input tissue of interest.
-
24.
Incubate on ice for 15 min
-
25.
Add 1 mL of MACS Buffer to samples.
-
26.
Centrifuge: 10 min, 300 × g, 4°C
-
27.
While samples are spinning, set up sorting columns in tissue culture hood (Figure 6)
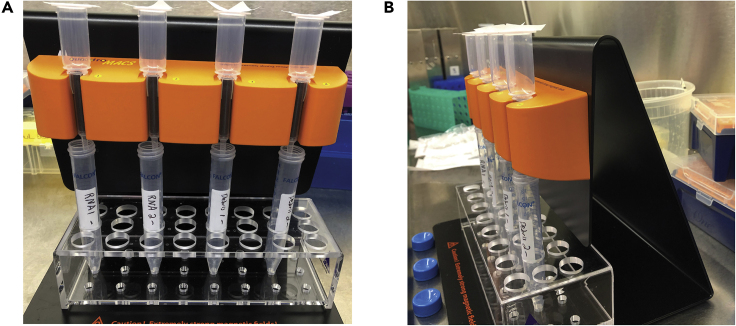
Figure 6.
Setup of LS Columns on MACS MultiStand.
(A) LS Columns are placed in the MACS MultiStand over opened 15 mL tubes. The LS Columns are topped with fresh nylon filters. (B) Ensure that LS Columns are placed tightly in the MACS MultiStand.
CRITICAL: Place the columns tightly in the magnet (must be fully in place). Then place the “-” tubes that will be used to collect CD11b negative cells under the columns. Top the columns with fresh nylon filters using autoclaved forceps to maintain sterility.
-
28.
Prewash each column with 3 mL of MACS Buffer through nylon filter.
CRITICAL: Pre-wet each filter by adding 1 mL of HBSS (with calcium/magnesium) to the middle of the filter and then drag to side.
-
29.
Remove samples from centrifuge and aspirate the supernatant. Resuspend pellets in 500 μL of MACS Buffer.
-
30.
Add cell suspension to nylon filter, and then rinse the old 15 mL tube with 2 mL of MACS Buffer and add it to the nylon filter.
-
31.
Add 3 mL of MACS Buffer to column.
-
32.
Once liquid has finished flowing through the columns, remove the “-” tubes and place on ice.
-
33.
Remove the columns from the magnet, and place on top of “+” tubes that will be used to collect CD11b positive cells (Video S6).
-
34.
Using a serological pipet, add 5 mL of MACS Buffer to the top of the column, and then quickly plunge column as hard as possible using the supplied plunger. Repeat this (for a total of 10 mL volume) for each column. Then place “+” tubes on ice with the “-” tubes (Video S7).
Be sure to dab top of “-” tube on the bottom of the LS column prior to removal, as there will be a drop of liquid containing negative flowthrough cells.
-
35.
Centrifuge: 10 min, 300 × g, 4°C
-
36.
Aspirate the supernatant (make sure to remove all bubbles, especially those from the “+” tubes)
CRITICAL: At this point you will only see a robust pellet in the “-” tubes and will not likely see a pellet in the “+” 15 mL falcon tubes. Because you likely will not see the pellet by eye, be sure to only displace the tube once to one side or the other and leave in place while aspirating off the supernatant (see Troubleshooting). Leave a meniscus of liquid covering the bottom of the tube prior to washing the pellet in 1X PBS, to avoid cell loss.
-
37.
Resuspend pellet in 400 μL of ice-cold 1X PBS
CRITICAL: washing the pellet in 1X PBS does risk some cell loss at this point and is not necessary for certain downstream applications such as quantification of microglial cytokine production in response to stimuli such as lipopolysaccharide (LPS). Short-term culture/incubation of microglial cells in media with and without pro-inflammatory stimuli can be accomplished by resuspending the pellet directly in your desired media at this point.
Plunge column as quickly and with as much force as possible.7
Washing the pellet in 1X PBS is necessary for downstream applications that could be affected by components of the MACS buffer (most notably the EDTA) such as single-cell RNA sequencing or flow cytometry.
-
38.
Transfer the 400 μL of cell resuspension to fresh 1.5 mL centrifuge tubes, rinse the old 15 mL tube with 800 μL of 1X PBS, and add this rinse to the new 1.5 mL tube as well.
-
39.
Centrifuge: 10 min, 300 × g, 4°C.
CRITICAL: Make sure to place all centrifuge tubes in the same orientation, as pellets may be difficult to visualize. See Troubleshooting for more detail on minimizing cell loss here.
-
40.
Carefully remove supernatant and resuspend in media of choice.
The media of choice will differ depending upon the intended usage for these isolated cells: (i) we use a DMEM-based media for cell culture and single-cell RNA sequencing applications, (ii) we resuspend cells in Trizol for RNA extraction applications, and (iii) we resuspend in PBS for flow cytometry applications. Final choice of media should be optimized in individual laboratories for their intended applications.
EXPECTED OUTCOMES
Using the above method, we have obtained microglial cell counts in the range of 100,000–200,000 cells per embryonic day 17.5 mouse brain, 100,000–300,000 per embryonic day 17.5 mouse placenta, and similar cell counts of 50,000–200,000 cells from bilateral hippocampus and prefrontal cortex and of 200,000–600,000 cells from whole forebrain of postnatal day 30–60 mice (Figure 7). Notably, the Percoll gradient isolation method permits isolation of sufficient microglia for downstream applications from individual embryonic mouse brains - with single cell sequencing
Figure 7.
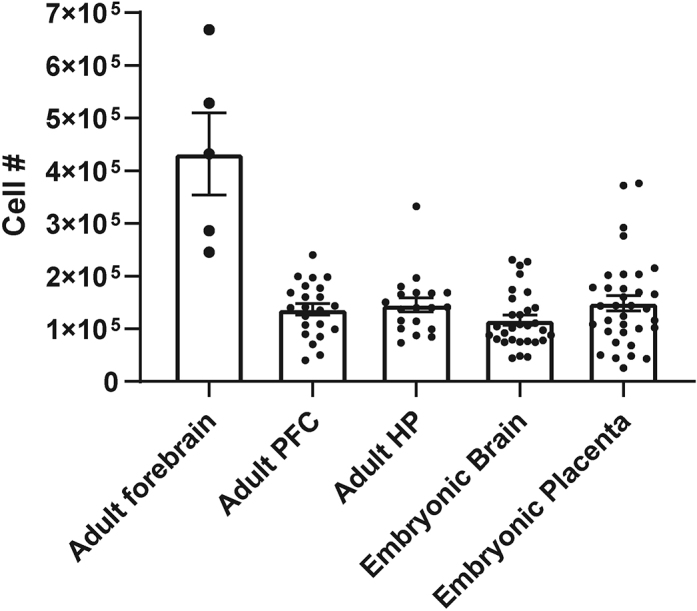
Cell Yields
Cell counts were obtained manually on a hemocytometer for CD11b+ cells isolated from adult forebrain, adult prefrontal cortex (PFC), adult hippocampus (HP), embryonic mouse forebrain, and embryonic mouse placenta.
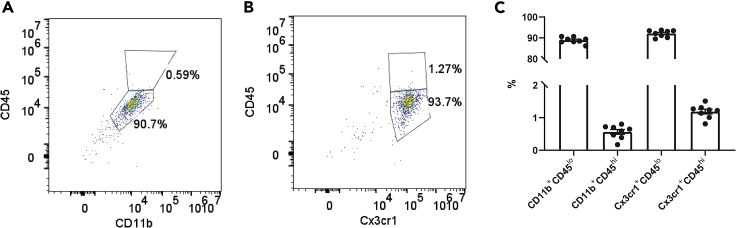
This method isolates all CD11b+ cells, which in the healthy brain are almost entirely microglia. We show in adult hippocampus that a high proportion of isolated cells are indeed brain microglia . After magnetic bead isolation, flow cytometry sorting for cells that were positive for either CD11b or Cx3cr1 were assessed for expression of CD45 (Figure 8). This protocol results in ~90% cells that are positive for either Cx3cr1 or CD11b, and under 1% of cells that are CD11b+CD45hi or Cx3cr1+CD45hi. CD45hi cells are often considered to be infiltrating macrophages, indicating a relative lack of non-microglial CD11b+ cells in our prep. However, it is the case that CD11b cell sorting may lead to non-parenchymal myeloid cell contamination (see ‘LIMITATIONS- Microglia vs other CD11b+ cells’ for further discussion).
Figure 8.
CD11b+ Magnetic Bead Isolation Pulls down Brain Microglia
(A and B) Gating strategy for (A) CD11b+ and CD45hi/lo and (B) Cx3cr1+ CD45hi/lo cells. Cells were isolated using CD11b+ magnetic bead isolation method presented in this protocol, and assessed for infiltration of non-parenchymal myeloid cells as assessed by increase in CD45 levels.
(C) Quantification of CD11b+CD45lo, CD11b+CD45hi, Cx3cr1+CD45lo, and Cx3cr1+CD45hi cells obtained using this protocol.
This method can be used to isolate cells at different developmental ages, as we demonstrated in (Hanamsagar et al.,2017). Bulk RNA sequencing analyses of microglia obtained using this method demonstrated that the microglial transcriptome matures with age, and at a different pace in male vs female microglia.
CD11b+ cells are not limited to the brain, and this methodology can be used to isolate CD11b+ cells from other tissues. For example, we have used this method to successfully isolate CD11b+ cells from individual mouse placentas, as well.
We have also successfully used this method to isolate microglia from fresh frozen human brain tissue. Successful isolation was confirmed by immunoblot positivity for Iba1 and negativity for GFAP and NeuN (Figure 9). Importantly, the protocol used to obtain CD11b+ cells from mouse brain tissue was unaltered in order to isolate CD11b+ cells from postmortem human tissue, beginning from Step 2. Step 2 mentions that human tissue was chopped with a razor blade and placed in enzyme digestion mix, after which human tissue underwent the exact same manipulations as mouse tissue. Our laboratory only cultures freshly-isolated cells, and as human postmortem tissue was all previously frozen, we have not attempted to culture CD11b+ cells from this tissue.
Figure 9.
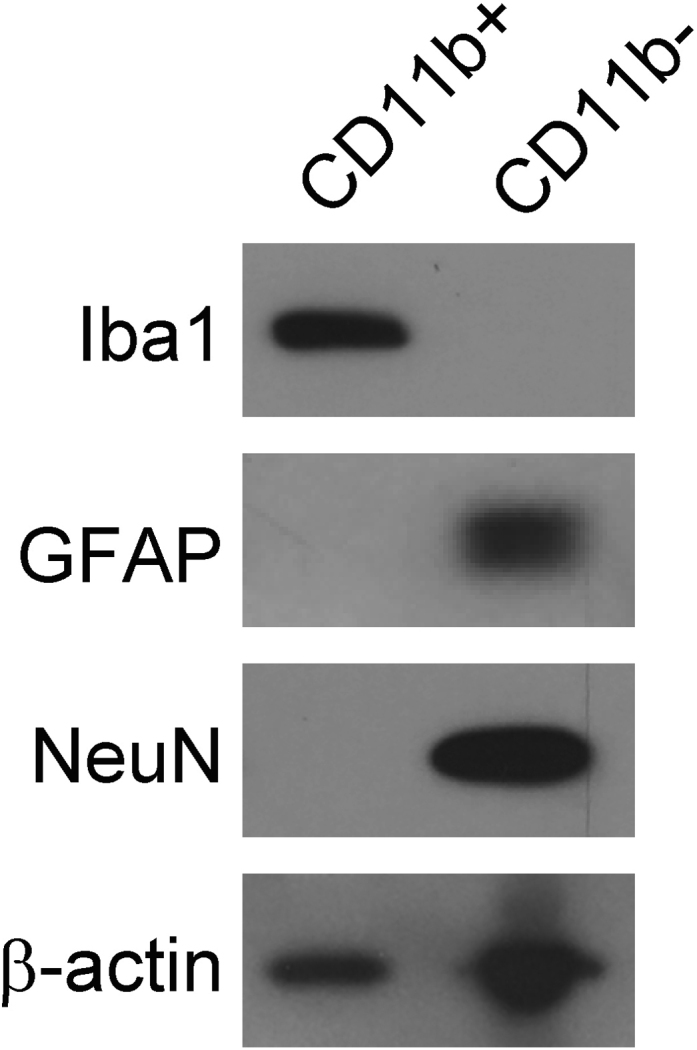
Lysates from Human CD11b+ or CD11b- Cells Were Run on Immunoblot and Probed for Iba1 (Microglia), GFAP (Astrocytes), and NeuN (Neurons)
CD11b+ cells were positive for Iba1 and negative for GFAP and NeuN, whereas CD11b- cells were negative for Iba1 and positive for GFAP and NeuN.
When culturing CD11b+ cells, we often treat and collect cells within 24 hr. However, we have been able to successfully culture these isolated cells for up to one week following isolation. We have not attempted to culture these cells longer-term, and this application would need to be optimized prior to use.
LIMITATIONS
Warm versus Cold Tissue Dissociation
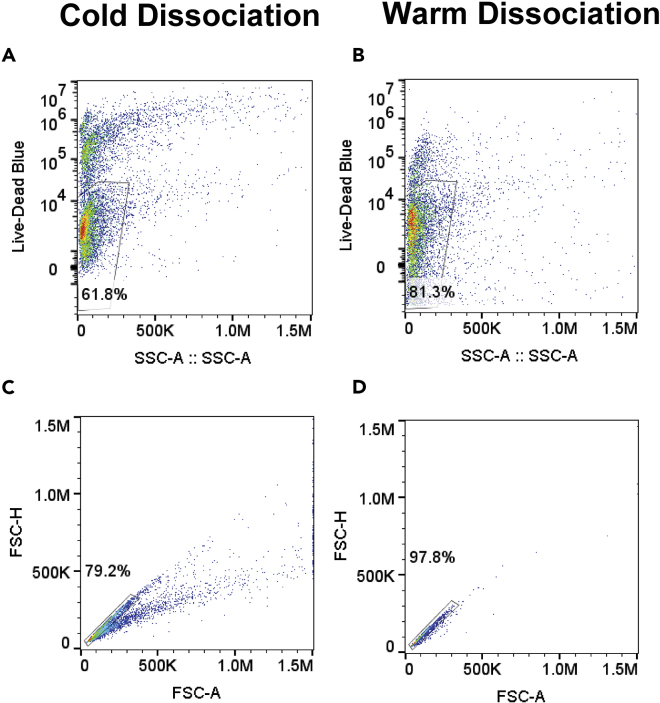
Although our lab has tried to implement microglia isolations using mechanical dissociation and maintaining cells at 4°C, we have encountered several limitations using this technique. First, despite many attempts, we are unable to attain a comparable yield between cold dissociations and warm enzymatic digestions. Warm enzymatic digestion still results in 2–3 times as many cells compared to cold mechanical dissociation. Second, when using panning methods to isolate microglia, removing microglia cells from the dish is difficult and the force required to remove the cells can negatively impact cell health. Third, cell viability is compromised limiting the ability to perform single cell sequencing. The tools/kits to eliminate dead cells either do not perform well or further reduce yield, thereby limiting an already small output. Lastly, cells maintained at 4°C appear less healthy (Figure 10). When centrifuging cells, each centrifugation from cold microglia yields more cellular debris and results in more clumping when producing single cell suspensions. In sum, microglia isolations using mechanical dissociation at 4°C, produce smaller yields with lower viability, and the cells are less healthy and more likely to clump, reducing the ability to perform state of the art single cell analysis (Figure 10).
Figure 10.
Cold Dissociation Results in Lowered Cell Viability and Fewer Single Cells than Does Warm Dissociation
(A and B) Gating strategy for Live-Dead Blue staining from (A) cold dissociation isolated CD11b+ cells and (B) warm dissociation isolated CD11b+ cells.
(C and D) Gating strategy for determining single cells from (C) cold dissociation isolated CD11b+ cells and (D) warm dissociation isolated CD11b+ cells.
Use of Embryonic Mouse Tissue
In our hands, the Miltenyi Debris Removal Solution does not work well for embryonic mouse tissue. When the Debris Removal Solution was attempted for embryonic mouse brain tissue, after the spin in Step 12/at Step 13, no cell layer was visualized in the gradients. Although we moved forward with the workflow and resuspended the pellet, ultimately very few to no cells were recovered. If the Debris Removal Solution is primarily targeted toward myelin removal, the relative lack of myelin in embryonic mouse tissue might explain why the solution is not optimal for an embryonic mouse tissue application. We therefore recommend the use of Percoll gradients for embryonic mouse tissue and have reliable cell yield with this method.
Microglia versus Other CD11b+ Cells
Although there are numerous methods used to isolate microglia from the brain, the method of dissociation and isolation can have a profound effect upon the function of these highly dynamic cells. For instance, fluorescence activated cell sorting (FACS) can upregulate immediate early gene expression (Ayata et al., 2018, Haimon et al., 2018, Li et al., 2019), and has been found to induce transcription of markers classically associated with the microglial proinflammatory state (Ayata et al., 2018), while methods such as RiboTag that use magnetic bead separation do not have similar gene alterations (Ayata et al., 2018, Haimon et al., 2018). FACS sorting has also been shown to alter the cellular redox state and metabolome in sorted cells (Llufrio et al., 2018). Perhaps the most enticing reason to utilize FACS for microglial isolation is the ability to further distinguish microglia from infiltrating immune cells. Transcardiac saline perfusion will eliminate any circulating CD11b+ cells, but any CD11b+ cells that are not microglia but are residing in the brain will be selected. A potential solution to this limitation could be to utilize the Miltenyi Multisort kits. For instance, the CD11b microbeads used in step 15 (Miltenyi #130-093-634) could be replaced by APC-tagged CD11b beads (Miltenyi #130-109-364), and the protocol for the Anti-APC MultiSort Kit (Miltenyi #130–091-255) could be followed. Central microglia vs peripheral immune cells could then be further sorted by a secondary antibody such as Cx3cr1. For instance, PE-tagged Cx3cr1 antibody (Biolegend #149005) coupled to anti-PE beads (Miltenyi #130-048-801) could be utilized to magnetically sort cells with CD11b and Cx3cr1. Alternatively, other secondary antibodies could be used to selectively isolate cells of interest without the confounds of FACS.
Protein Mass Spectrometry Using Post-mortem Human Brain Tissue
We were able to successfully isolate microglia from human post-mortem brain regions using this method, as confirmed by positivity for Iba1 and negativity for GFAP and NeuN by immunoblot (see Figure 9). However, users may struggle to successfully perform proteomic analyses in isolated CD11b+ cells using this method. Two main limitations that we encountered during these studies were (a) the relatively low protein abundance in the isolated CD11b+ cells obtained from 300 mg of human brain tissue and (b) the high levels of bovine serum albumin (BSA) contamination within the sample. The source of the BSA in our samples is the MACS Buffer (1%, see above in described recipe) used during CD11b bead incubation and LS column steps. BSA, a protein widely used for routine experiments is known to be a common contaminant for proteomics due to both its molecular size and its abundance (Hodge et al., 2013, Bell et al., 2009, Pellitteri-Hahn et al., 2006). In general, high levels of protein contamination can cause the loss of low abundance proteins given the large peaks from highly abundant proteins with similar size (Hodge et al., 2013). In our experimental conditions, the presence of large amounts of BSA (up to 90% of the intensity measured in the raw MS data) masked the protein peaks during Mass Spec analysis. Thus, the low protein abundance of CD11b+ cells together with the presence of BSA as a contaminant of the sample negatively affected proteomics analysis. Optimization of this protocol, perhaps with further wash steps to further rid sample of BSA, would be necessary before performing protein mass spectrometry analyses.
Use of Fetal Bovine Serum
We use 5% fetal bovine serum (FBS) in our enzyme digestion mix. Serum has been demonstrated to activate microglia in vitro, as serum-exposed microglia exhibit less ramified morphologies and significantly altered gene expressions (Bohlen et al., 2017, Bohlen et al., 2019). Although we do include FBS in our enzyme digestion mix, our previously published data using this method indicate negligible activation of classical proinflammatory cytokines. For instance, isolated microglia treated with vehicle in vitro demonstrated low levels of IL-1b and TNF-a which was significantly stimulated by treatment with LPS (Rivera et al., 2019), and embryonic mouse brain and placental CD11b+ cells showed significant stimulation of TNF-a release following LPS challenge (Edlow et al., 2019). ELISA run on cultured CD11b+ and CD11b- cells isolated from embryonic mouse tissue show low levels of TNF-α release without LPS stimulation, and a significant stimulation of TNF-α production following LPS treatment (Figure 11). Although our data indicates negligible activation of microglia from the FBS contained in our enzyme digestion mix, the presence of FBS must be considered as a potential limitation of this method.
Figure 11.
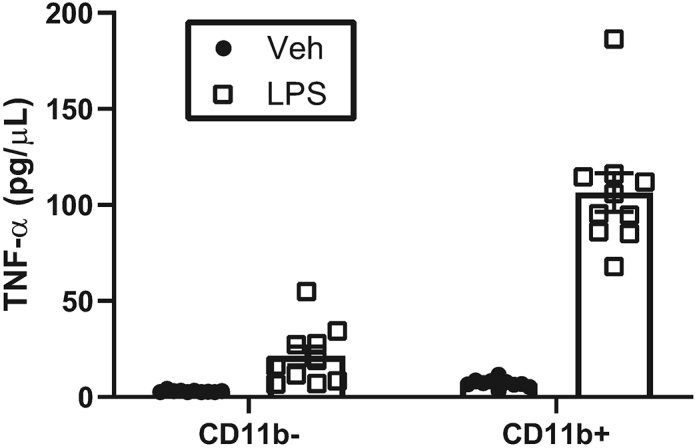
Low Level of Basal Cytokine Release by Placental CD11b+ Cells
Protein concentrations for TNF-α released into culture media by CD11b- or CD11b+ cells following 2 hr lipopolysachharide (LPS) or vehicle (Veh) stimulation were determined by ELISA.
TROUBLESHOOTING
Problem 1
70% Percoll underlay does not drain out of the unflamed Pasteur pipet tip.
Potential Solution
Gently lift the tip of the unflamed Pasteur pipet from the bottom of the falcon tube and place back down. Doing this several times should allow the air bubbles to escape from the bottom of the tip, and the 70% Percoll will subsequently drain.
Problem 2
Difficulty visualizing the interphase layer at step 13 while pipetting (critical for pipetting out the microglia, which will be the only cellular material moving forward from this point).
Potential Solution
We recommend placing the tube such that the interphase layer is at eye level. The layer is most easily visualized before any cells are removed. Cells should be removed by pipetting in circles just above the 30/70 gradient. If 70% Percoll from below the interphase layer is accidentally obtained, a second gradient after the subsequent spin (step 14) may form. If this happens, see Problem 4 and Solution.
Problem 3
Thin or non-robust interphase cell layer in the Percoll gradients.
Potential Solution
We have found that the robustness of Percoll gradients and the cell yield following Step 10 is sensitive to specific centrifuges and spin times. A centrifuge with swinging baskets is critical for Step 10, as is the use of zero brake and slow acceleration. The spin, acceleration, and brake settings are optimized for the Eppendorf 5810R centrifuge with A-4-81 rotor and swinging bucket attachments, and may need to be altered for other centrifuge/rotors.
Problem 4
A second Percoll gradient is visualized after the spin in step 14.
Potential Solution
If this occurs, a band of cells will be visible near the bottom of the tube above the pellet which contains the majority of your cells for subsequent steps. This occurs when 70% Percoll was included in the 4 mLs of the interphase cell layer. This can be avoided by errring on the side of staying above 70% Percoll when pipetting out the interphase layer. To minimize cell loss if this second gradient does occur, we harvest the cells in this second gradient. To do this, aspirate the supernatant to just above the visible cell layer (typically several mm above the pellet), and pipet out the second interphase layer in a circular motion as described in step 12 (only one time). Then aspirate off the remainder of the supernatant above the pellet (may help to tilt tube at this point), and add the cells from the second interphase to the pellet before moving forward. For example of layer removal, see Video S8.
Supernatant is aspirated, cell layer is sucked into pipet and saved, lower supernatant is aspirated, and cells are placed back with other pelleted cells.
Problem 5
Lower than expected cell yield.
Potential Solution
We have identified the following points at which unexpected cell loss can occur:
-
1)
Step 10 if using Percoll gradients (see above regarding Solution for thin/non-robust interphase layer after the 20 min spin)
-
2)
Step 6: If the filter is not pre-wet properly and the pre-wet area extends over the entire surface of the filter, the added cell suspension can track around the top of the falcon tube and cell loss can occur.
-
3)
Steps 19–24: If performing the ‘Molecular Protocol’ that does not utilize Percoll or Debris Removal Solution, it is possible that large amounts of tissue from older animals can cause the column to clog up from excess myelination. While we have not experienced this using tissue from postnatal day 0 to postnatal day 60 mice, using tissue from older animals may cause this issue. If this occurs, we suggest utilization of either Percoll or Debris Removal Solution protocols.
-
4)
Step 28 and Step 32: At these point s when the pellet can be difficult to visualize, care must be taken when aspirating supernatants to leave sufficient supernatant covering the bottom of the tube if pellet is unclear, and to keep the falcon or eppendorf tube slightly tilted to the same side throughout aspiration to avoid disturbing the pellet.
Acknowledgments
We thank Haley Norris for technical assistance. This work was supported by NIEHS R01 ES025549 to S.D.B., NIMH F32 MH116604 to E.A.B., and NICHD R01-HD100022 to A.G.E.
AUTHOR CONTRIBUTIONS
E.A.B. and S.D.B. conceived the manuscript. E.A.B., C.L.B., T.P., G.S-V., C.J.S., A.G.E., and S.D.B. wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100035.
Contributor Information
Evan A. Bordt, Email: ebordt@mgh.harvard.edu.
Staci D. Bilbo, Email: staci.bilbo@duke.edu.
References
- Ayata P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018;21:1049–1060. doi: 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A.W., Deutsch E.W., Au C.E., Kearney R.E., Beavis R., Sechi S., Nilsson T., Bergeron J.J. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J., Bennett F.C., Tucker A.F., Collins H.Y., Mulinyawe S.B., Barres B.A. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron. 2017;94:759–773.e8. doi: 10.1016/j.neuron.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J., Bennett F.C., Bennett M.L. Isolation and culture of microglia. Curr. Protoc. Immunol. 2019;125:e70. doi: 10.1002/cpim.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlow A.G., Glass R.M., Smith C.J., Tran P.K., James K., Bilbo S. Placental macrophages: a window into fetal microglial function in maternal obesity. Int. J. Dev. Neurosci. 2019;77:60–68. doi: 10.1016/j.ijdevneu.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimon Z., Volaski A., Orthgiess J., Boura-Halfon S., Varol D., Shemer A., Yona S., Zuckerman B., David E., Chappell-Maor L. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018;19:636–644. doi: 10.1038/s41590-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R., Alter M.D., Block C.S., Sullivan H., Bolton J.L., Bilbo S.D. Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia. 2017;65:1504–1520. doi: 10.1002/glia.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge K., Have S.T., Hutton L., Lamond A.I. Cleaning up the masses: exclusion lists to reduce contamination with HPLC-MS/MS. J. Proteomics. 2013;88:92–103. doi: 10.1016/j.jprot.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Cheng Z., Zhou L., Darmanis S., Neff N.F., Okamoto J., Gulati G., Bennett M.L., Sun L.O., Clarke L.E. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–223.e10. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llufrio E.M., Wang L., Naser F.J., Patti G.J. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018;16:381–387. doi: 10.1016/j.redox.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellitteri-Hahn M.C., Warren M.C., Didier D.N., Winkler E.L., Mirza S.P., Greene A.S., Olivier M. Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J. Proteome Res. 2006;5:2861–2864. doi: 10.1021/pr060287k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera P.D., Hanamsagar R., Kan M.J., Tran P.K., Stewart D., Jo Y.C., Gunn M., Bilbo S.D. Removal of microglial-specific MyD88 signaling alters dentate gyrus doublecortin and enhances opioid addiction-like behaviors. Brain Behav. Immun. 2019;76:104–115. doi: 10.1016/j.bbi.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pipet aggregated tissue until all tissue moves freely up into the Pasteur pipet. Then pipet up and down 20 times.
Begin with tube tilted at 45° angle and slowly layer 1X PBS on top of Debris REMOVAL Solution layer. As more 1X PBS is added, slowly bring tube back to a vertical position.
Use finger to cover Pasteur pipet as it is placed under the 30% Percoll. Slowly pipet 70% Percoll into the Pasteur pipet and let it release under the 30% Percoll layer. Cover pipet with finger and gently remove.
Be sure to dab top of “-” tube on the bottom of the LS column prior to removal, as there will be a drop of liquid containing negative flowthrough cells.
Plunge column as quickly and with as much force as possible.7
Supernatant is aspirated, cell layer is sucked into pipet and saved, lower supernatant is aspirated, and cells are placed back with other pelleted cells.