Extended Data Fig. 6 |.
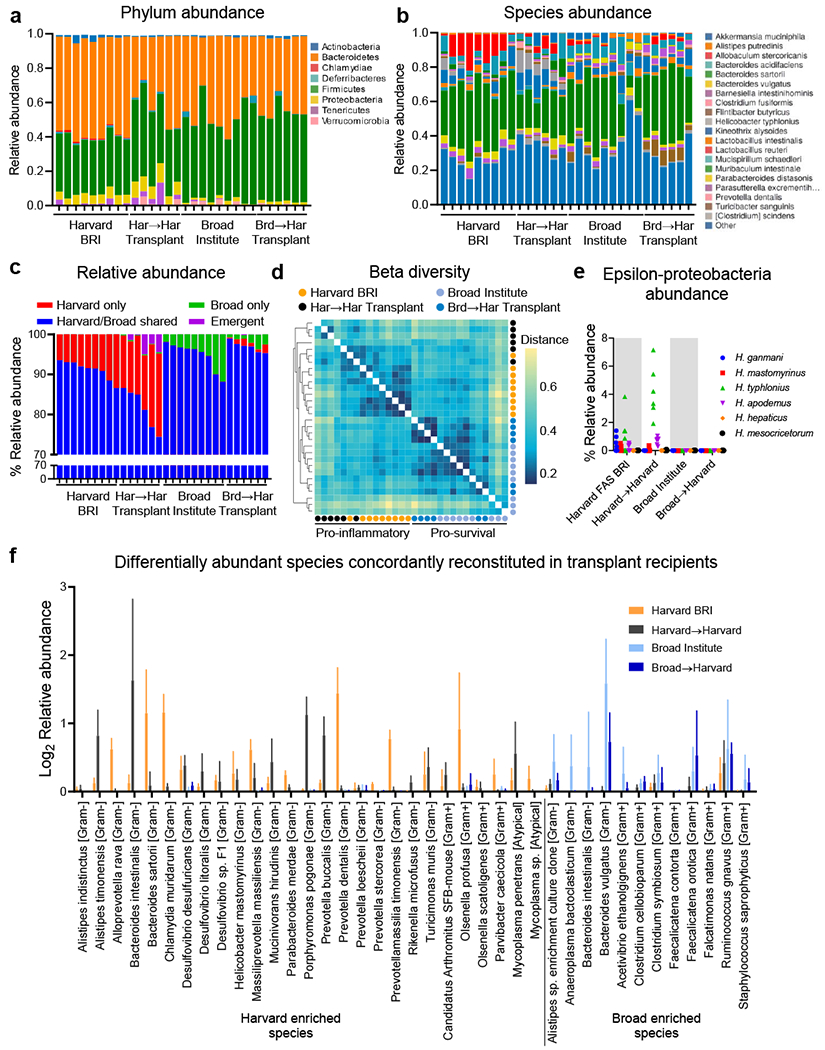
Environment enriched bacteria engraft fecal transplant recipients
a-f, Analysis of bacteria in feces 10-weeks-post-transplant from mice in Fig 3e by 16S rDNA sequencing. Each bar represents a fecal sample from an individual cage, a, Phylum level and b, species level relative abundance, c, Relative abundance of bacterial species grouped as those only observed in cages from Harvard BRI (Harvard-only), those only observed in cages from the Broad Institute (Broad-only), those observed in cages from Harvard BRI and the Broad Institute (Harvard/Broad-shared) or those not observed in Harvard BRI or Broad Institute cages but detectable in transplant recipient cages (Emergent), d, Bray-Curtis dissimilarity matrix of feces beta diversity, e, Relative abundance of epsilon proteobacteria [Helicobacter], (f) Putative pro-inflammatory species (n=27) enriched in pro-inflammatory environments (Harvard BRI/JHU) that were also enriched in Harvard->Harvard recipients and putative pro-survival species (n=12) enriched in pro-survival environments (Broad/Jackson Labs) and enriched in Broad->Harvard recipients.
