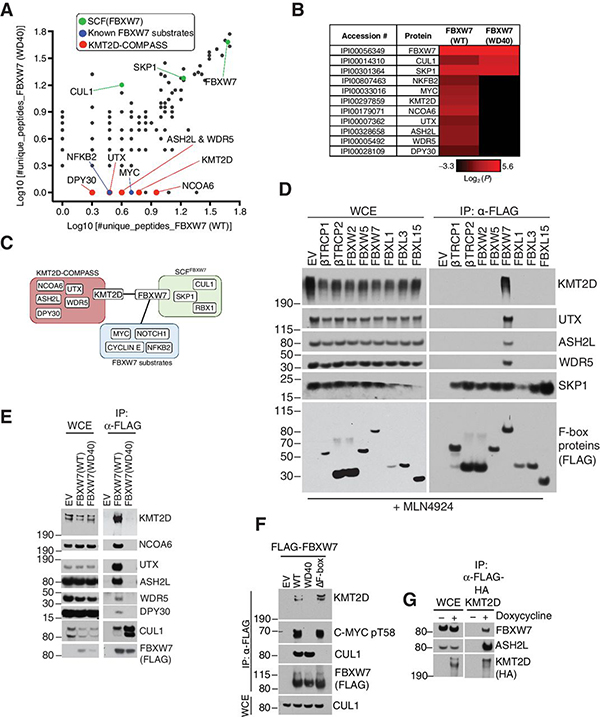
Figure 1. Proteomics-based approach identifies KMT2D as a substrate-like interactor of FBXW7.
A, Scatter Plot of log10(#unique peptides+1) identified in the mass spectrometry analysis of FBXW7 immunoprecipitates. Known SCF interactors (green), known substrates (blue), and putative substrates (red) are highlighted. B, Heatmap of log2(#unique peptides+0.1) for the proteomic in (a.) C, Schematic representation of SCF-FBXW7 substrate interaction network. D, Immunoblot of indicated proteins from whole cell extracts (WCE) or α-FLAG immunoprecipitates (FLAG-IP) from HEK293T cells overexpressing the indicated FLAG-F-box proteins. Treatment with MLN4924 was for 6 hours. E, Immunoblot of indicated proteins from WCE or FLAG-IP samples from HEK293T cells overexpressing the indicated FLAG-FBXW7 constructs. F, Immunoblot of indicated proteins from FLAG-IP samples from HEK293T cells overexpressing the indicated FLAG-sFBXW7 constructs. G, Immunoblot of the indicated proteins from WCE or FLAG-IP samples in BJAB cells. FLAG-HA-KMT2D was stably expressed in BJAB DLBCL cells using the doxycycline-inducible Flp-In™ T-Rex™ System (doxycycline treatment: 1 μg/mL for 16 hours).