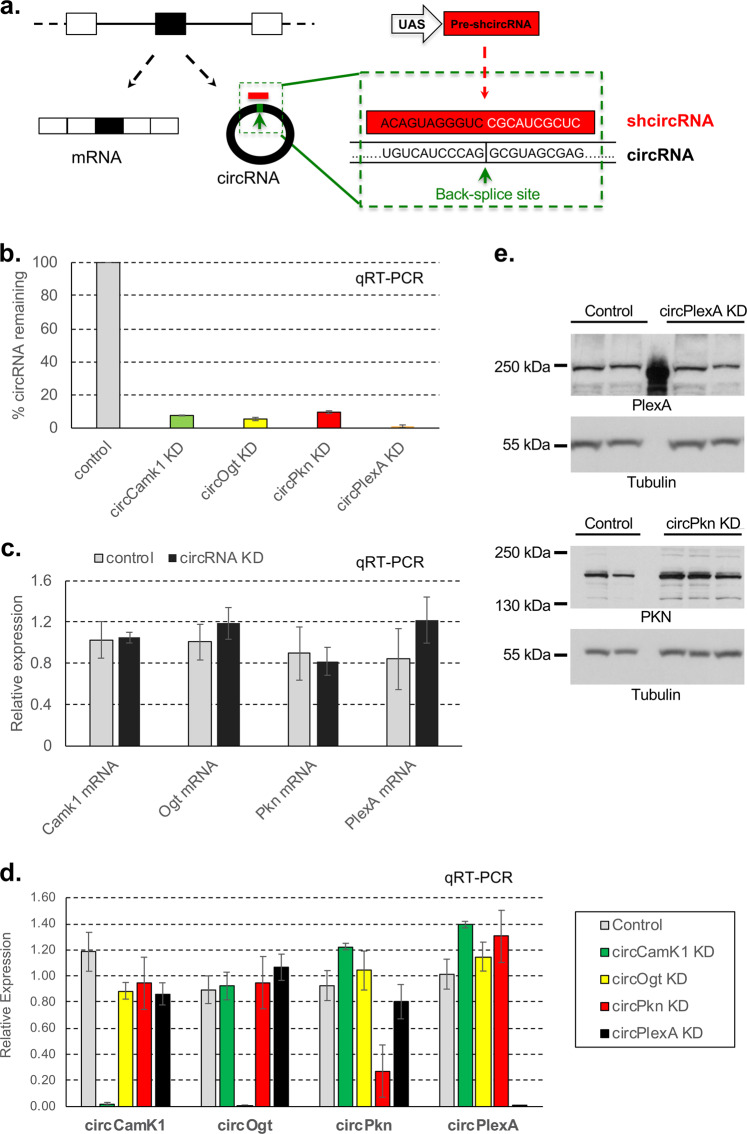
Fig. 1. circRNAs can be specifically downregulated by genetically encoded shRNA in vivo.
a Scheme of the shRNA strategy to knockdown circRNAs in vivo. ShRNAs are generated from a UAS-based miR-1 like precursor (see Materials and methods). b Relative levels of the targeted circRNAs in fly heads expressed as percentage of control (actin-Gal4 flies). Levels of the indicated circRNAs were assess by qRT-PCR using the rp49 mRNA as normalization control (n = 3, error bars represent standard error of the mean). c Relative expression levels of the indicated mRNAs in the four knockdown strains. Levels were normalized to mean expression in control line (actin-Gal4 flies). In all cases we utilized fly heads as source of material and assessed the levels of the mRNAs by qRT-PCR using rp49 as normalization control (n = 3, error bars represent standard error of the mean). d Levels of the indicated circRNAs in the four knockdown strains. In all cases we utilized fly heads as source of material and assessed the levels of the circRNAs by qRT-PCR using rp49 as normalization control (n = 3, error bars represent standard error of the mean). e Levels of the PKN and PLEXA proteins (and Tubulin as loading control) as assayed by western blot in heads of control (actin-Gal4), circPkn and circPlexA KD strains.