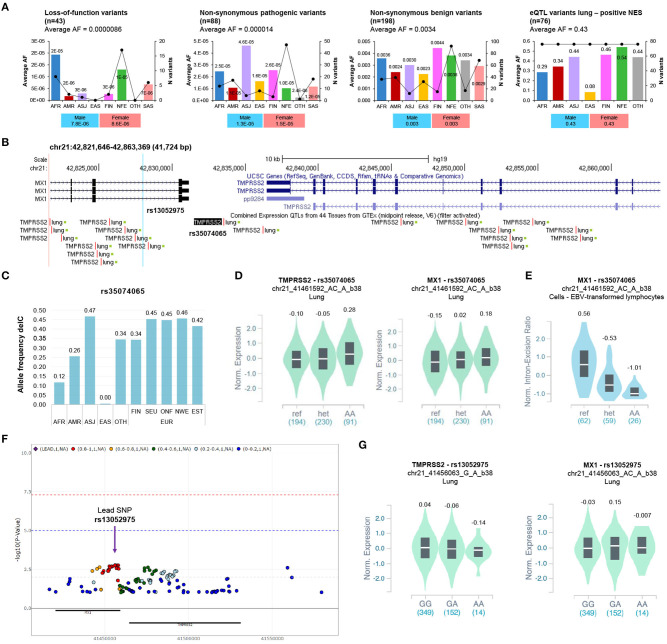
Figure 1.
Analysis of the coding variants and the eQTLs of TMPRSS2 locus. (A) The alternative allele frequency (AF) distribution of non-synonymous pathogenic, benign, loss-of-function, and eQTL-lung variants (positive NES) of TMPRSS2 in different populations. The Y axis of each bar plot shows average allele frequency (AF) in each population. The second right axis displays the number of variants within each population. The bar graph below shows the AF distribution in the overall gnomAD population stratified according to gender. AFR, African/African American; AMR, Latino/Admixed American; ASJ, Ashkenazi Jewish; EAS, East Asian; FIN, Finnish; NFE, Non-Finnish European; SAS, South Asian; OTH, Other (population not assigned). (B) Schematics of the genomic region encompassing eQTL lung variants of TMPRSS2 locus (NES positive ≥ 0.1) by Genome Browser (GRCh37/hg19, https://genome.ucsc.edu/). The most significant eQTL-lung (rs35074065) and the eQTL rs13052975, associated with severe COVID-19, are highlighted. (C) The allele frequencies of delC variant rs35074065 were annotated by the gnomAD database (WGS data). AFR, African/African American; AMR, Latino/Admixed American; ASJ, Ashkenazi Jewish; EAS, East Asian; OTH, Other (population not assigned); FIN, Finnish; SEU, Southern European; ONF, Other non-Finnish European; NEW, North-Western European; EST, Estonian. (D) Violin plot showing the effect of the eQTL rs35074065 on TMPRSS2 and MX1 expression in lung (TMPRSS2: p = 3.9e-11; NES = 0.13; MX1: p = 0.000010; NES = 0.20). (E) Violin plot showing the effect of the sQTL rs35074065 on MX1 splicing isoform expression (p = 1.6e-13; NES = −0.83). (F) Regional association plot (P < 0.1) of variants at TMPRSS2 locus (proximity: ± 50Kb; analysis II, adjusted for top 10 PCs, age, sex) and respiratory failure in COVID-19 (data from COVID-19 GWAS results browser, https://ikmb.shinyapps.io/COVID-19_GWAS_Browser/). (G) Violin plot showing the effect of the eQTL rs13052975 on TMPRSS2 and MX1 expression in lung (TMPRSS2: P = 0.0063; NES = −0.072; MX1: P = 0.15; NES = −0.090).